Chapter 2 Data Searches and Pairwise Alignments 20040308





- Slides: 47

Chapter 2 Data Searches and Pairwise Alignments 暨南大學資訊 程學系 黃光璿 2004/03/08 1

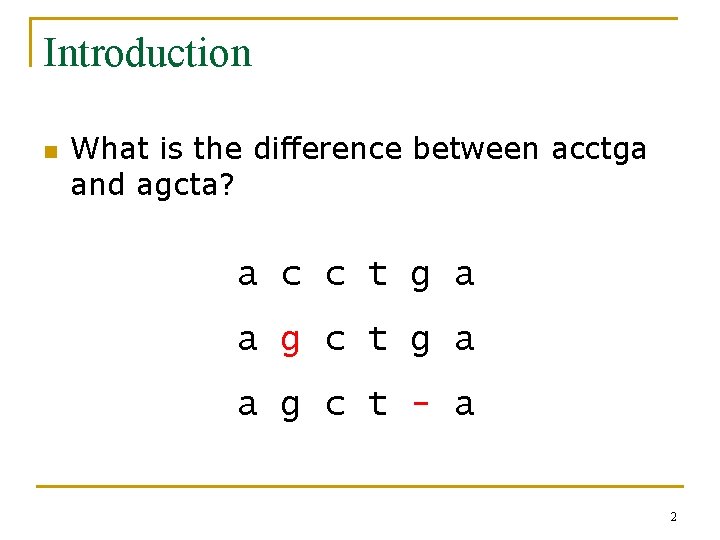
Introduction n What is the difference between acctga and agcta? a c c t g a a g c t - a 2

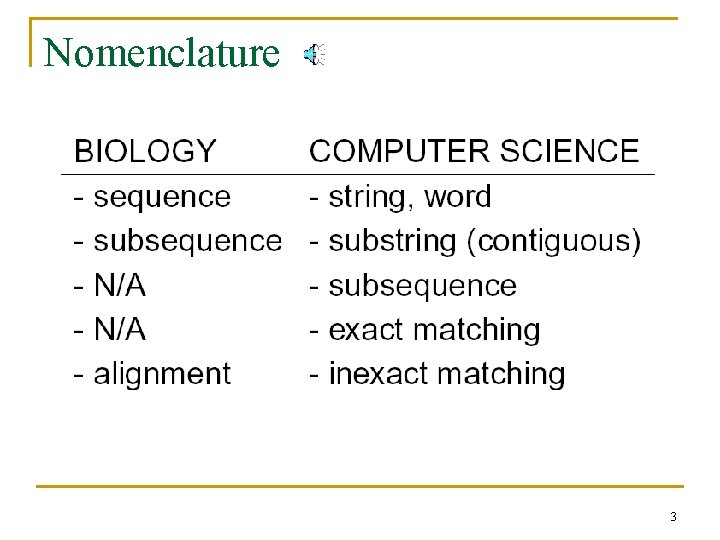
Nomenclature 3

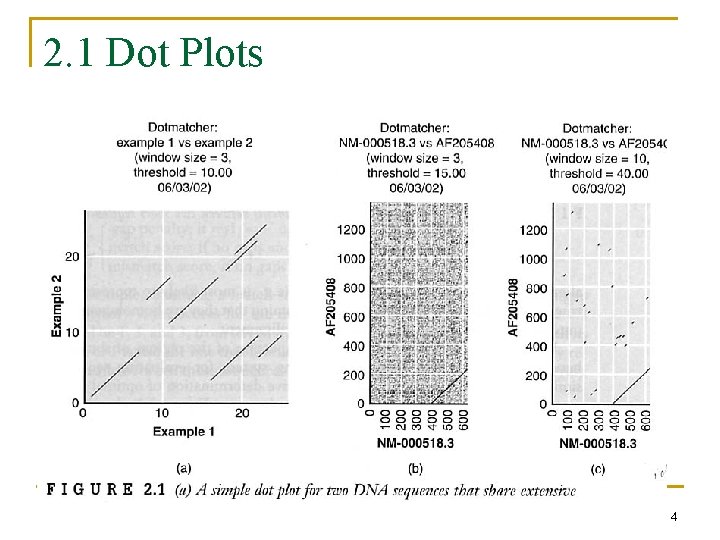
2. 1 Dot Plots 4

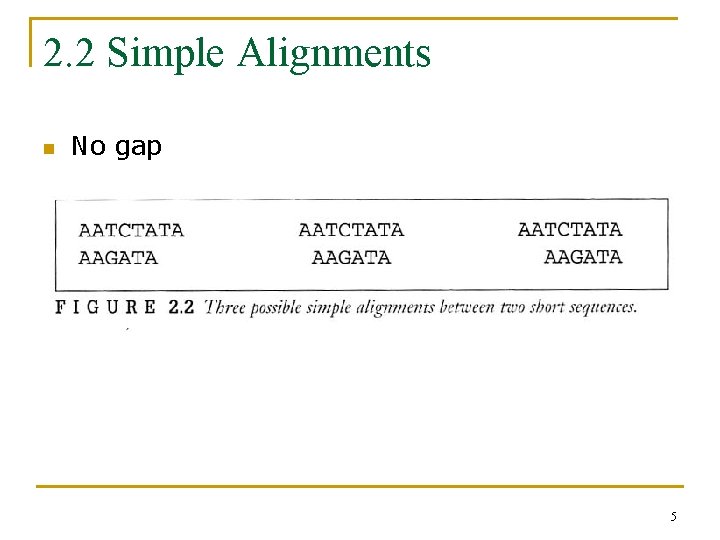
2. 2 Simple Alignments n No gap 5

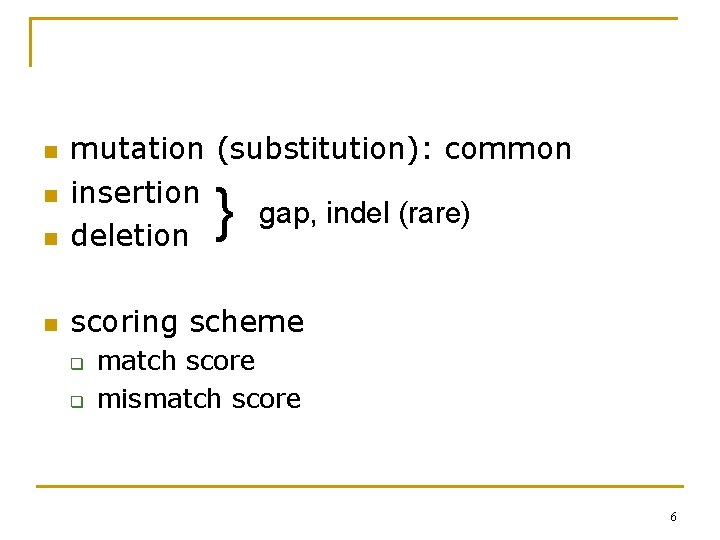
n mutation (substitution): common insertion gap, indel (rare) deletion n scoring scheme n n } q q match score mismatch score 6

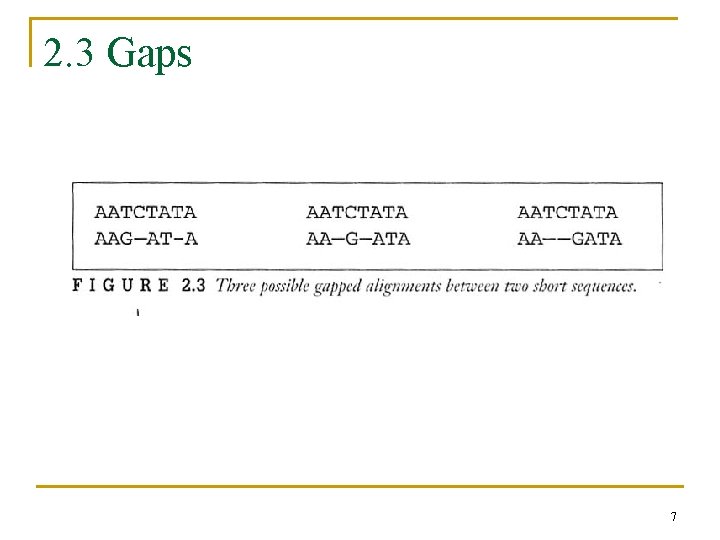
2. 3 Gaps 7

2. 3. 1 Gap Penalty n n uniform gap affine gap q q origination penalty length penalty 8

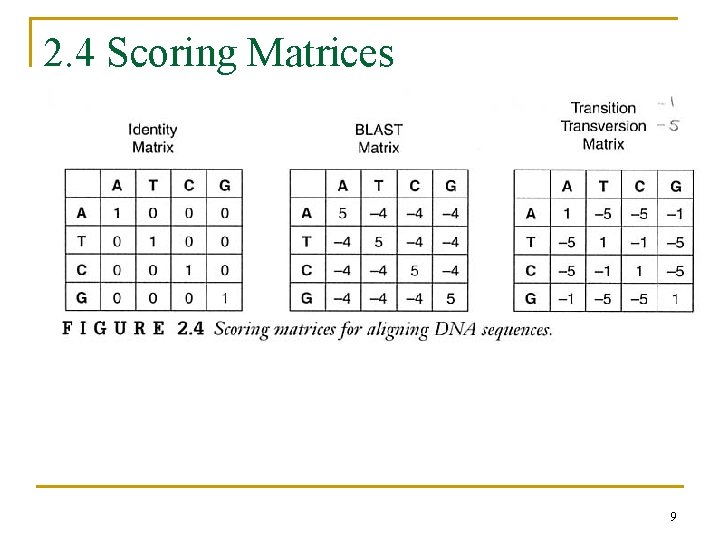
2. 4 Scoring Matrices 9


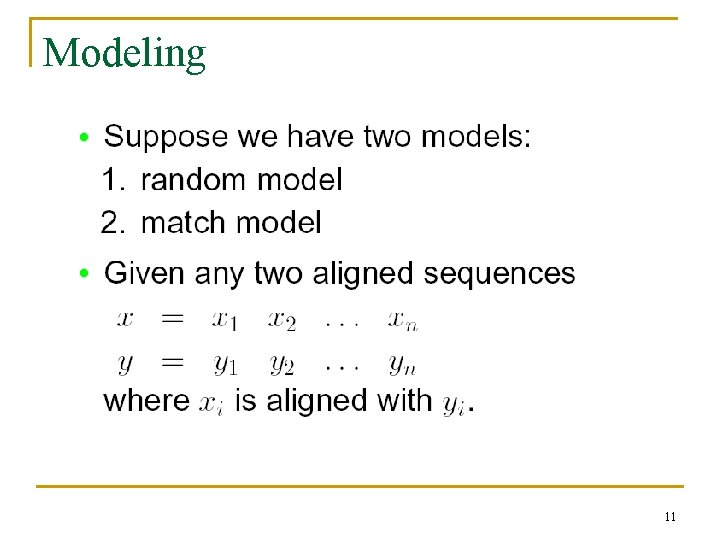
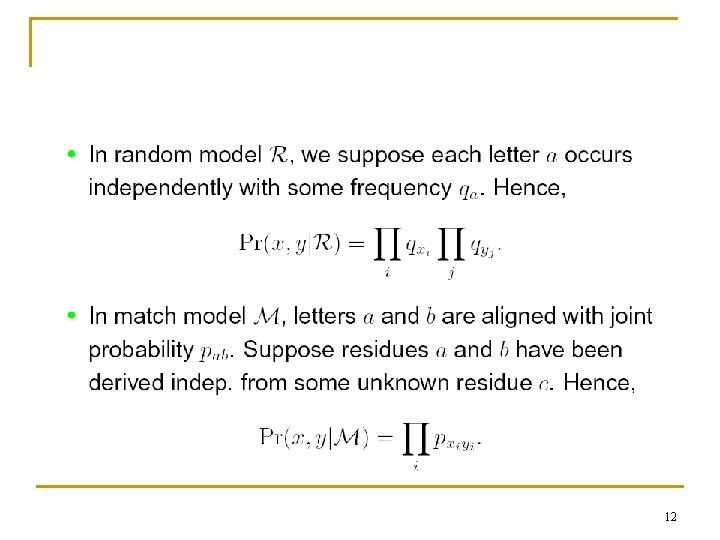
Modeling 11

12

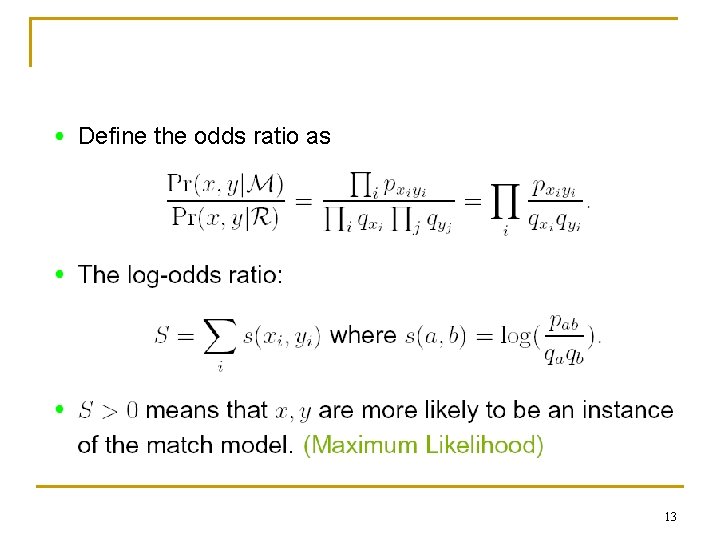
Define the odds ratio as 13

2. 4. 1 PAM Matrices n n Dayhoff, Schwartz, Orcutt (1978) Point Accepted Mutation q Based on observed substitution rates n q Input n q (Box. 2. 1) A set of observed substitution rates Output n PAM-1 matrix (log-odds matrix) 14

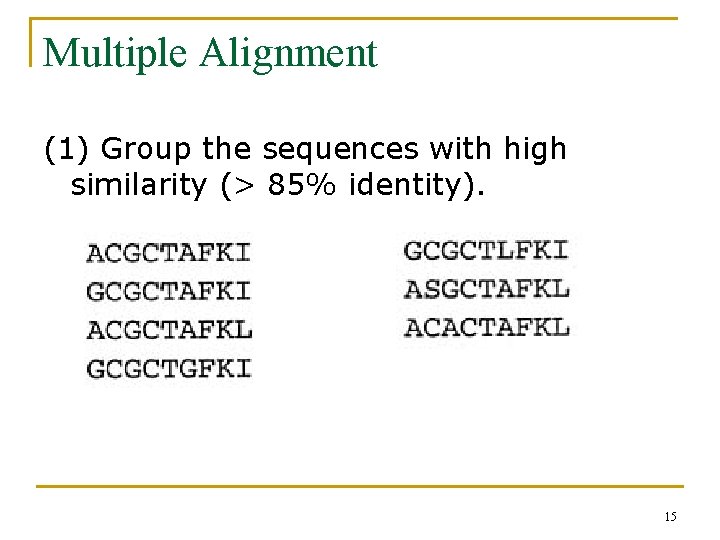
Multiple Alignment (1) Group the sequences with high similarity (> 85% identity). 15

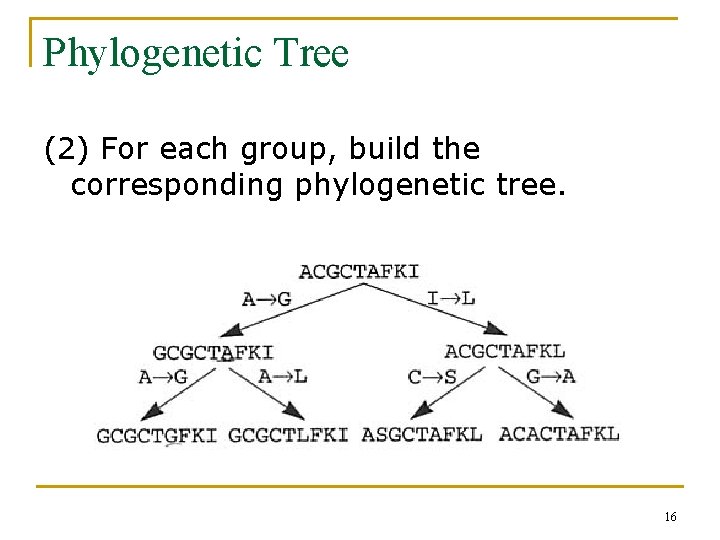
Phylogenetic Tree (2) For each group, build the corresponding phylogenetic tree. 16

Mutation Frequency A->G, I->L, A->G, A->L, C->S, G->A (3) FG, A=3 17

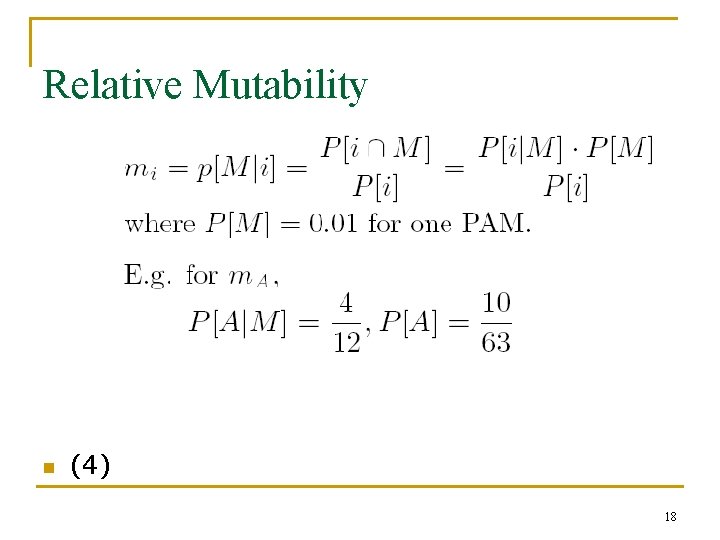
Relative Mutability n (4) 18

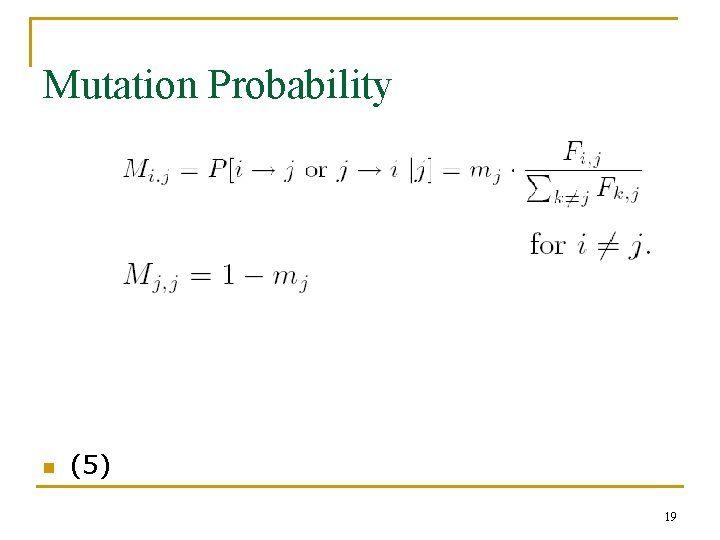
Mutation Probability n (5) 19

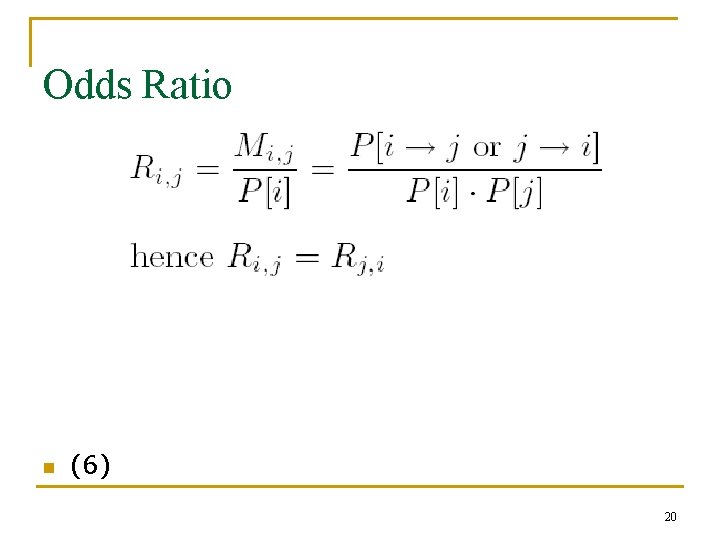
Odds Ratio n (6) 20

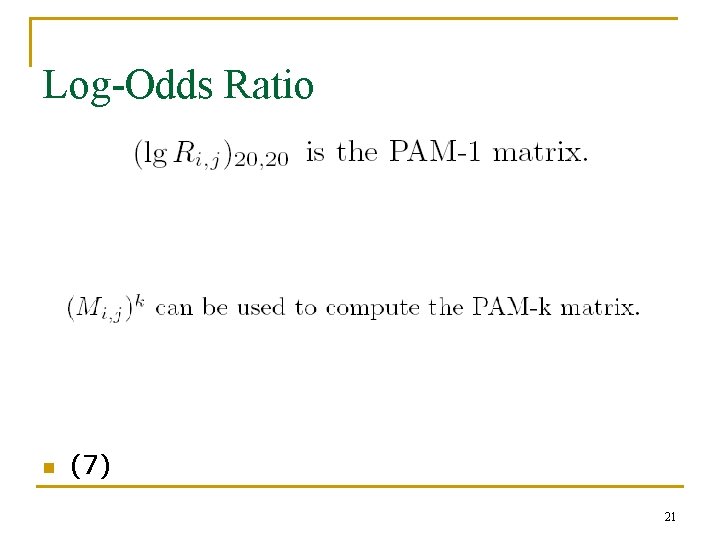
Log-Odds Ratio n (7) 21

n Which PAM matrix is the most appropriate? q q n n the length of the sequences How closely the sequences are believed to be related. PAM 120 for database search PAM 200 for comparing two specific proteins 22

2. 4. 2 BLOSUM Matrices n n n Henikoff & Henikoff (1992) PAM-k: k愈大, 愈不相似 BLOSUM-k: k愈大愈相似 BLOSUM 62: for ungapped matching BLOSUM 50: for gapped matching 23

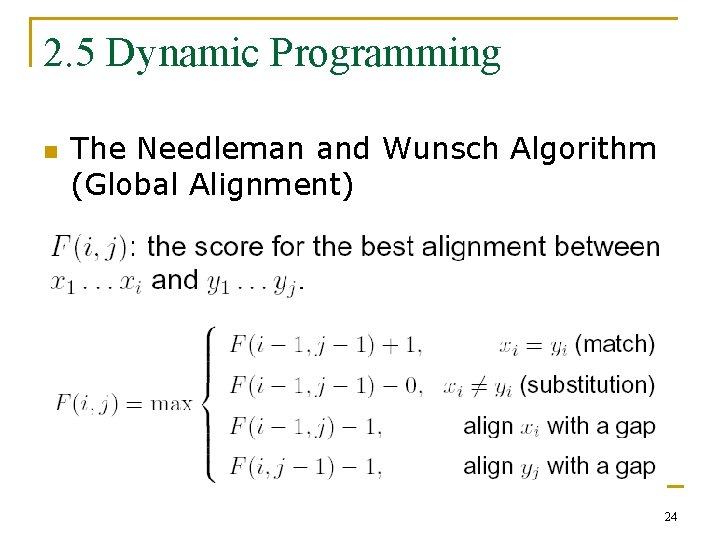
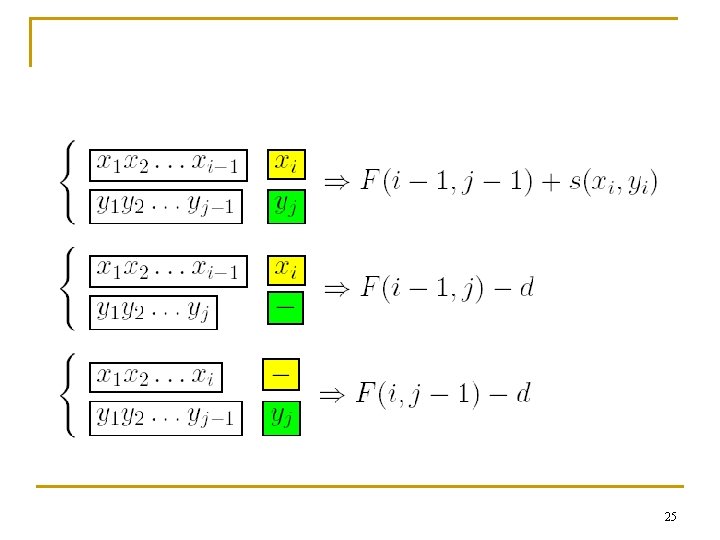
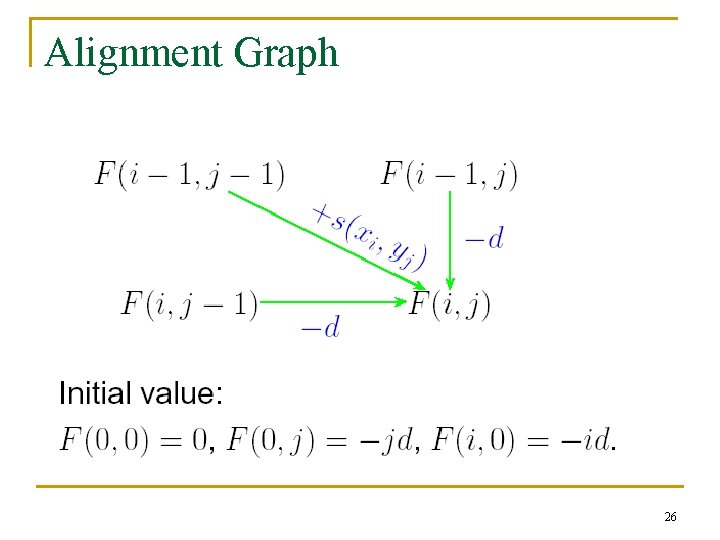
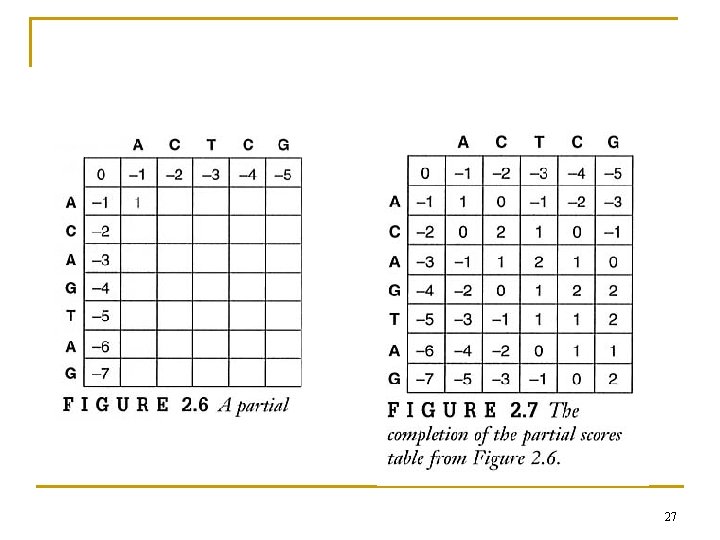
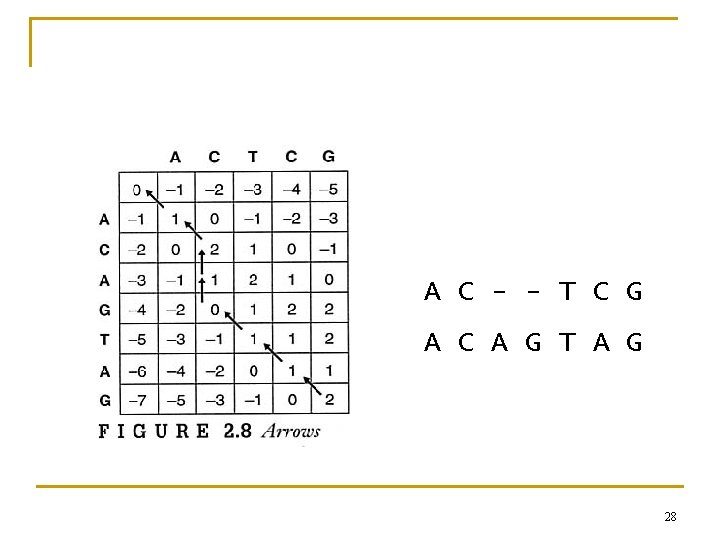
2. 5 Dynamic Programming n The Needleman and Wunsch Algorithm (Global Alignment) 24

25

Alignment Graph 26

27

A C - - T C G A C A G T A G 28

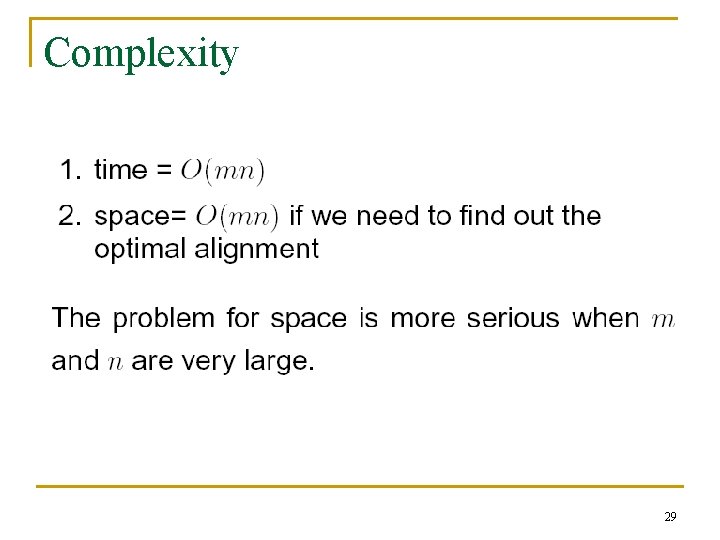
Complexity 29

2. 6 Global and Local Alignments n n Semi-global alignment Local alignment 30

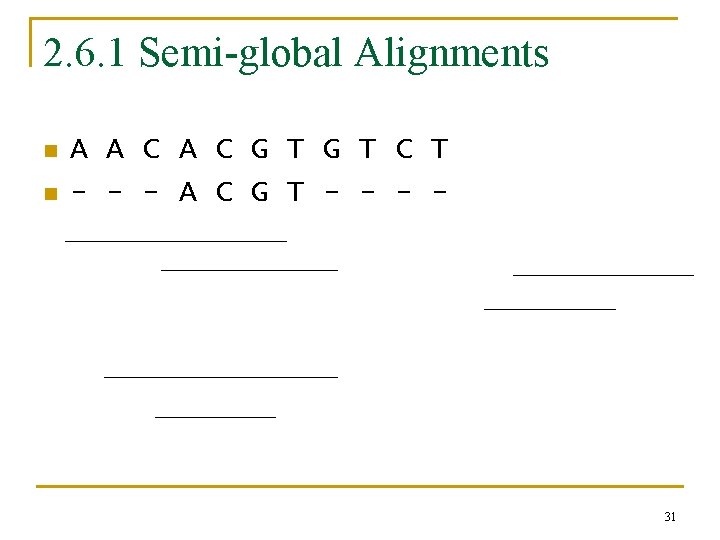
2. 6. 1 Semi-global Alignments n n A A C G T C T - - - A C G T - - 31

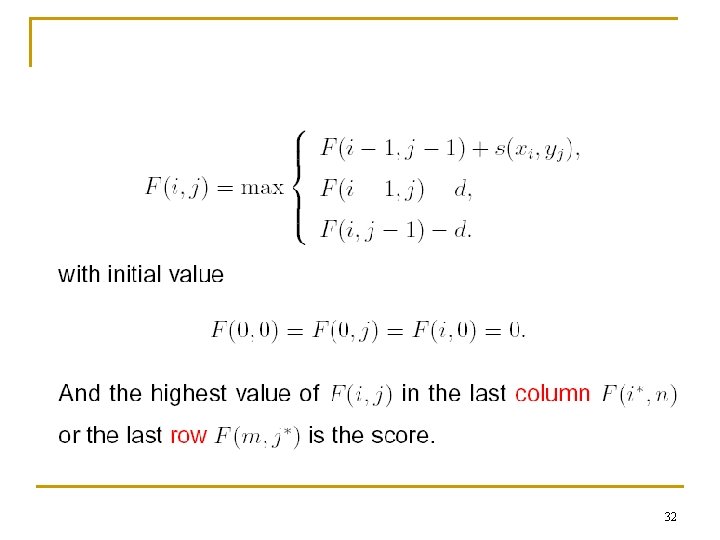
32

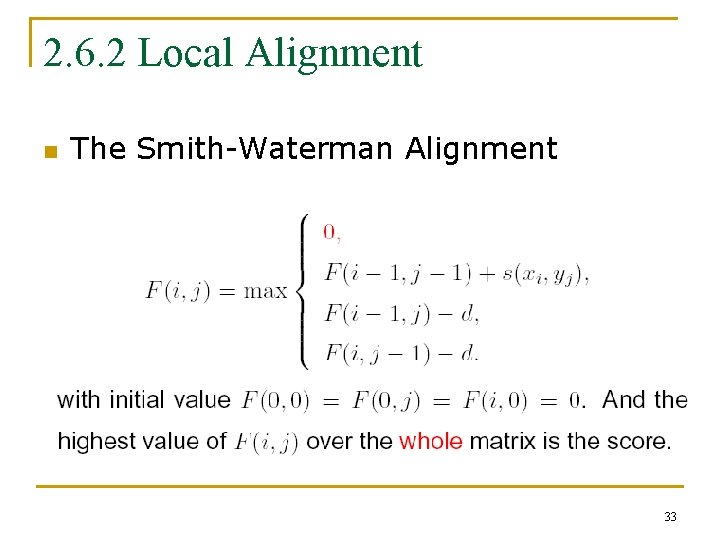
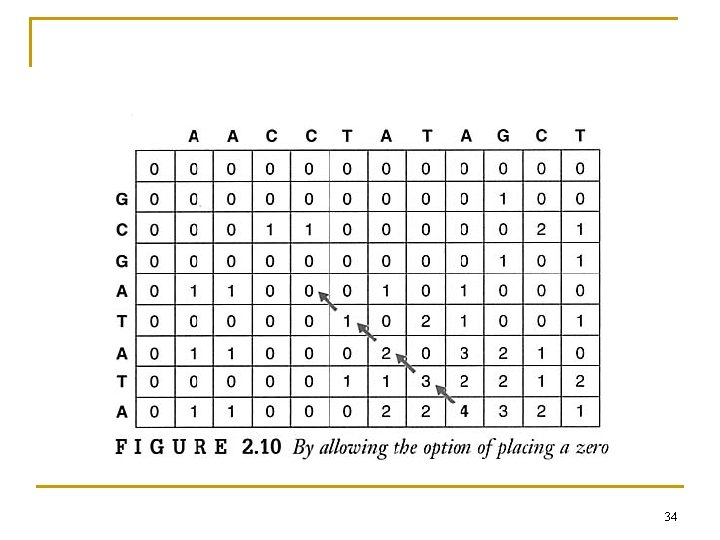
2. 6. 2 Local Alignment n The Smith-Waterman Alignment 33

34

2. 7 Database Searches n n BLAST and its relatives FASTA and related algorithms 35

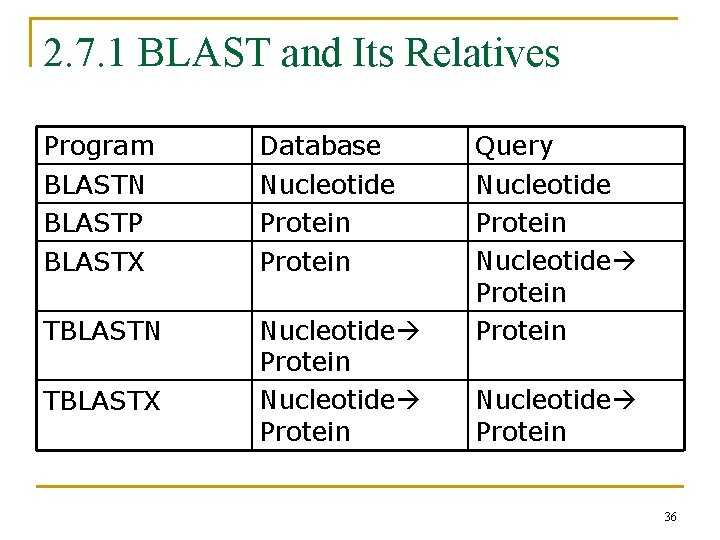
2. 7. 1 BLAST and Its Relatives Program BLASTN BLASTP BLASTX Database Nucleotide Protein TBLASTN Nucleotide Protein TBLASTX Query Nucleotide Protein Nucleotide Protein 36

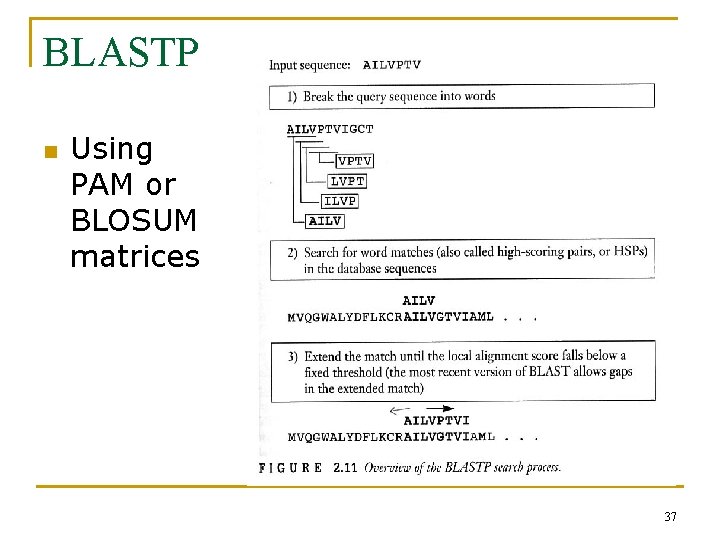
BLASTP n Using PAM or BLOSUM matrices 37

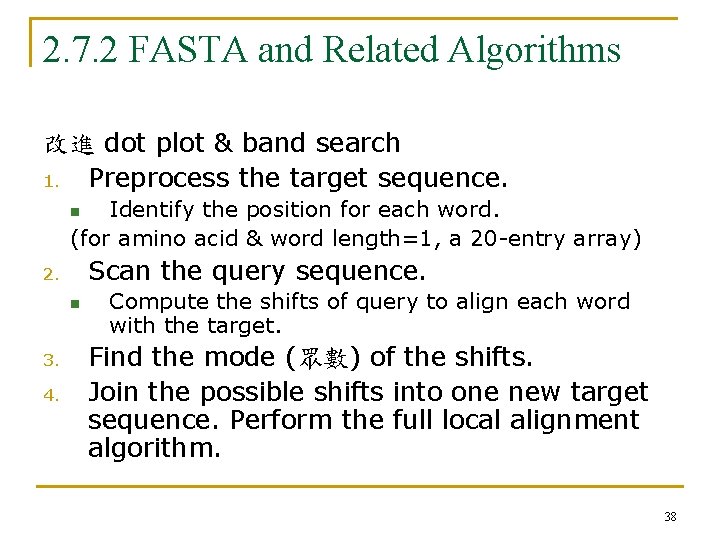
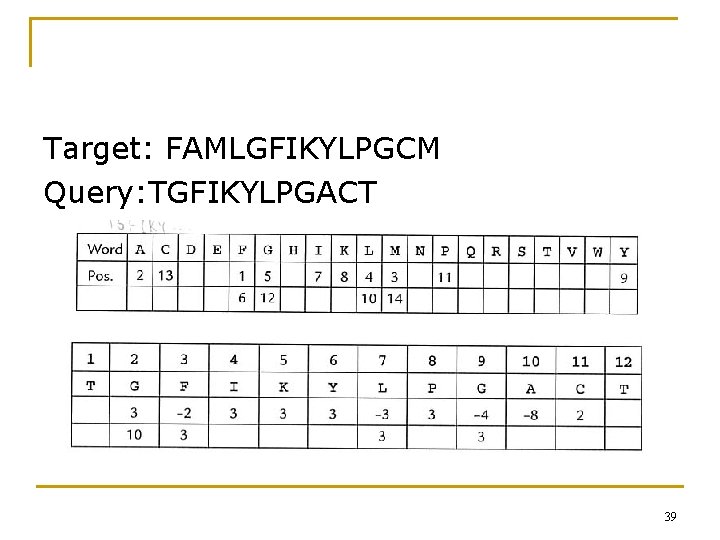
2. 7. 2 FASTA and Related Algorithms 改進 dot plot & band search 1. Preprocess the target sequence. Identify the position for each word. (for amino acid & word length=1, a 20 -entry array) n Scan the query sequence. 2. n 3. 4. Compute the shifts of query to align each word with the target. Find the mode (眾數) of the shifts. Join the possible shifts into one new target sequence. Perform the full local alignment algorithm. 38

Target: FAMLGFIKYLPGCM Query: TGFIKYLPGACT 39

2. 7. 3 Alignment Scores and Statistical Significance of Database Searches n related model v. s. random model q q q n S-score: the alignment score E-score: expected number of sequences with score >= S by random chance P-score: probability that one or more sequences with score >= S would be found randomly Low E & P are better. 40

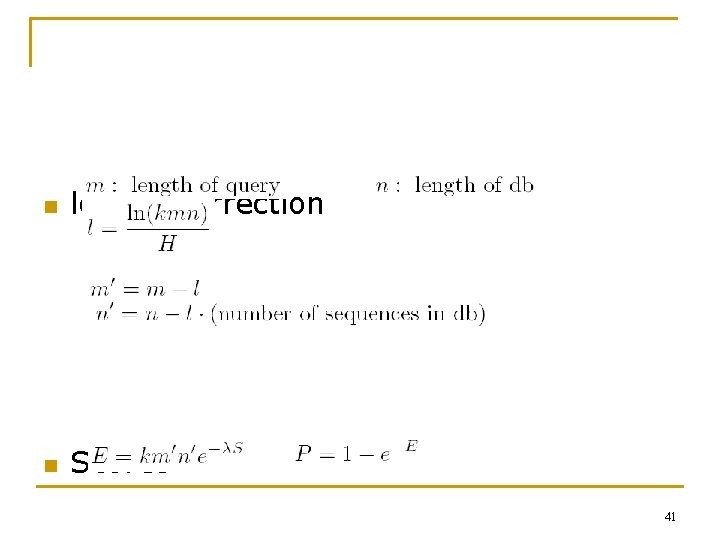
n length correction n Scores 41

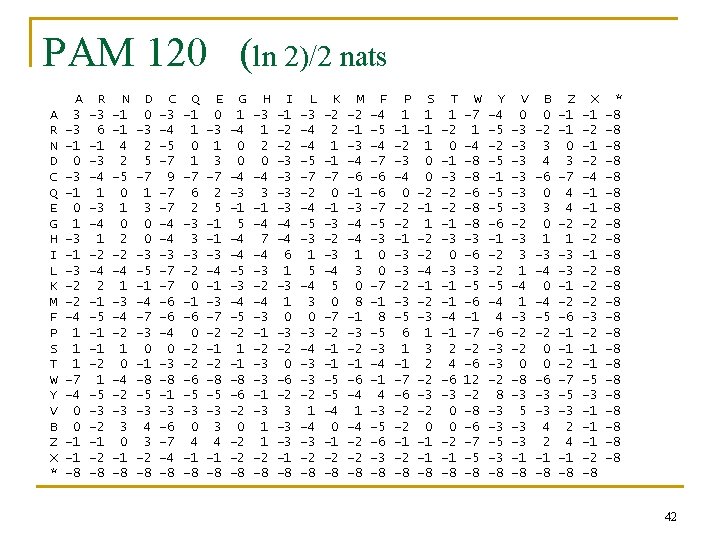
PAM 120 (ln 2)/2 nats A R N D C Q E G H I L K M F P S T W Y V B Z X * A 3 -3 -1 0 1 -3 -2 -2 -4 1 1 1 -7 -4 0 0 -1 -1 -8 R -3 6 -1 -3 -4 1 -2 -4 2 -1 -5 -1 -1 -2 1 -5 -3 -2 -1 -2 -8 N -1 -1 4 2 -5 0 1 0 2 -2 -4 1 -3 -4 -2 1 0 -4 -2 -3 3 0 -1 -8 D 0 -3 2 5 -7 1 3 0 0 -3 -5 -1 -4 -7 -3 0 -1 -8 -5 -3 4 3 -2 -8 C -3 -4 -5 -7 9 -7 -7 -4 -4 -3 -7 -7 -6 -6 -4 0 -3 -8 -1 -3 -6 -7 -4 -8 Q -1 1 0 1 -7 6 2 -3 3 -3 -2 0 -1 -6 0 -2 -2 -6 -5 -3 0 4 -1 -8 E 0 -3 1 3 -7 2 5 -1 -1 -3 -4 -1 -3 -7 -2 -1 -2 -8 -5 -3 3 4 -1 -8 G 1 -4 0 0 -4 -3 -1 5 -4 -4 -5 -3 -4 -5 -2 1 -1 -8 -6 -2 0 -2 -2 -8 H -3 1 2 0 -4 3 -1 -4 7 -4 -3 -2 -4 -3 -1 -2 -3 -3 -1 -3 1 1 -2 -8 I -1 -2 -2 -3 -3 -4 -4 6 1 -3 1 0 -3 -2 0 -6 -2 3 -3 -3 -1 -8 L -3 -4 -4 -5 -7 -2 -4 -5 -3 1 5 -4 3 0 -3 -4 -3 -3 -2 1 -4 -3 -2 -8 K -2 2 1 -1 -7 0 -1 -3 -2 -3 -4 5 0 -7 -2 -1 -1 -5 -5 -4 0 -1 -2 -8 M -2 -1 -3 -4 -6 -1 -3 -4 -4 1 3 0 8 -1 -3 -2 -1 -6 -4 1 -4 -2 -2 -8 F -4 -5 -4 -7 -6 -6 -7 -5 -3 0 0 -7 -1 8 -5 -3 -4 -1 4 -3 -5 -6 -3 -8 P 1 -1 -2 -3 -4 0 -2 -2 -1 -3 -3 -2 -3 -5 6 1 -1 -7 -6 -2 -2 -1 -2 -8 S 1 -1 1 0 0 -2 -1 1 -2 -2 -4 -1 -2 -3 1 3 2 -2 -3 -2 0 -1 -1 -8 T 1 -2 0 -1 -3 -2 -2 -1 -3 0 -3 -1 -1 -4 -1 2 4 -6 -3 0 0 -2 -1 -8 W -7 1 -4 -8 -8 -6 -8 -8 -3 -6 -3 -5 -6 -1 -7 -2 -6 12 -2 -8 -6 -7 -5 -8 Y -4 -5 -2 -5 -1 -5 -5 -6 -1 -2 -2 -5 -4 4 -6 -3 -3 -2 8 -3 -3 -5 -3 -8 V 0 -3 -3 -3 -2 -3 3 1 -4 1 -3 -2 -2 0 -8 -3 5 -3 -3 -1 -8 B 0 -2 3 4 -6 0 3 0 1 -3 -4 0 -4 -5 -2 0 0 -6 -3 -3 4 2 -1 -8 Z -1 -1 0 3 -7 4 4 -2 1 -3 -3 -1 -2 -6 -1 -1 -2 -7 -5 -3 2 4 -1 -8 X -1 -2 -4 -1 -1 -2 -2 -2 -3 -2 -1 -1 -5 -3 -1 -1 -1 -2 -8 * -8 -8 -8 -8 -8 -8 42

Applications n n Reconstructing long sequences of DNA from overlapping sequence fragments Determining physical and genetic maps from probe data under various experiment protocols Database searching Comparing two or more sequences for similarities 43

n n Protein structure prediction (building profiles) Comparing the same gene sequenced by two different labs 44

2. 8 Multiple Sequence Alignemnts n CLUSTAL q n R. G. Higgins & P. M. Sharp, 1988 CLUSTALW q q Sequences are weighted according to how divergent they are from the most closely related pair of sequences. Gaps are weighted for different sequences. 45

Summary n n notion of similarity the scoring system used to rank alignments the algorithms used to find optimal scoring alignment the statistical method used to evaluate the significance of an alignment score 46

參考資料及圖片出處 1. 2. 3. 4. Fundamental Concepts of Bioinformatics Dan E. Krane and Michael L. Raymer, Benjamin/Cummings, 2003. BLAST, by I. Korf, M. Yandell, J. Bedell, O‘Reilly & Associates, 2003. (天瓏代理) Biological Sequence Analysis – Probabilistic Models of Proteins and Nucleic Acids R. Durbin, S. Eddy, A. Krogh, and G. Mitchison, Cambridge University Press, 1998. Biochemistry, by J. M. Berg, J. L. Tymoczko, and L. Stryer, Fith Edition, 2001. 47