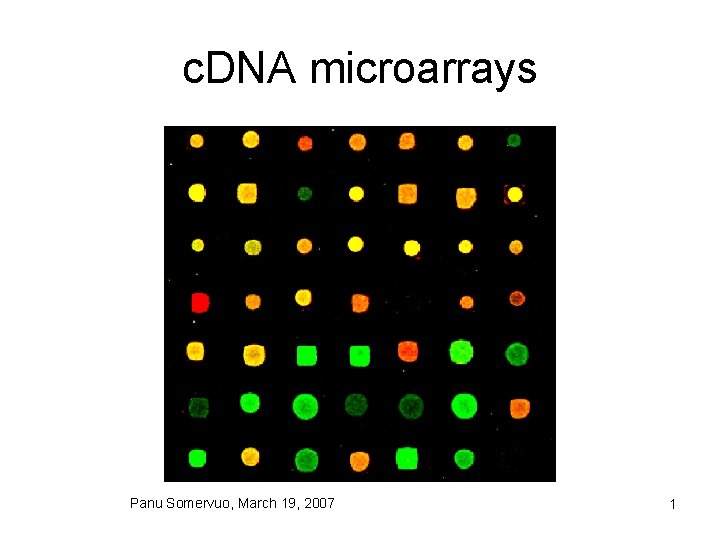
c DNA microarrays Panu Somervuo March 19 2007



- Slides: 19

c. DNA microarrays Panu Somervuo, March 19, 2007 1

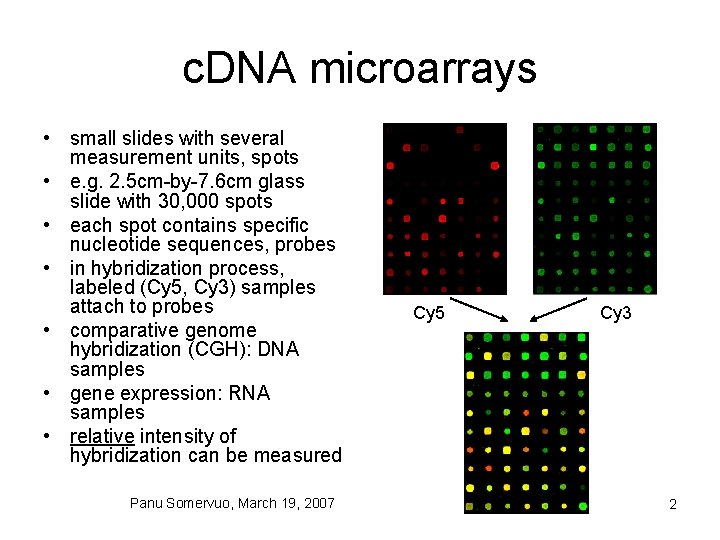
c. DNA microarrays • small slides with several measurement units, spots • e. g. 2. 5 cm-by-7. 6 cm glass slide with 30, 000 spots • each spot contains specific nucleotide sequences, probes • in hybridization process, labeled (Cy 5, Cy 3) samples attach to probes • comparative genome hybridization (CGH): DNA samples • gene expression: RNA samples • relative intensity of hybridization can be measured Panu Somervuo, March 19, 2007 Cy 5 Cy 3 2

Data flow • • • biological data, DNA/RNA extraction, fluoresence dye labeling, hybridization array scanning image processing: spot segmentation datafile • • data preprocessing and normalization: data analysis 1: statistical tests to find differentially expressed genes gene lists • data analysis 2: biological interpretations of results Panu Somervuo, March 19, 2007 3

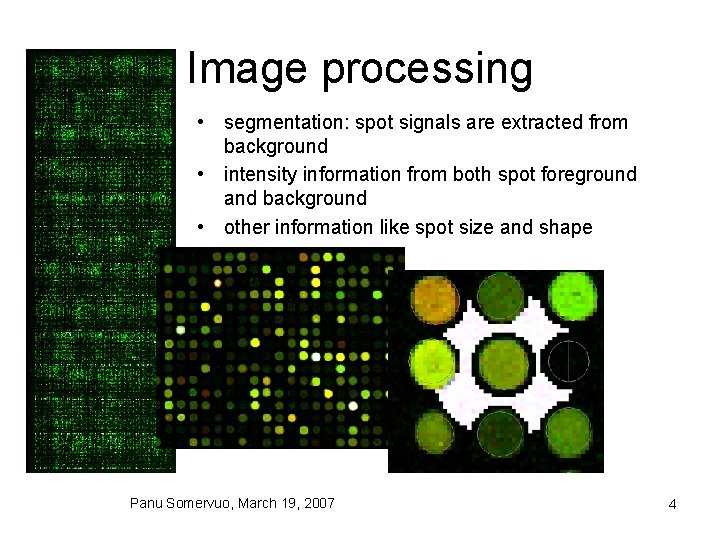
Image processing • segmentation: spot signals are extracted from background • intensity information from both spot foreground and background • other information like spot size and shape Panu Somervuo, March 19, 2007 4

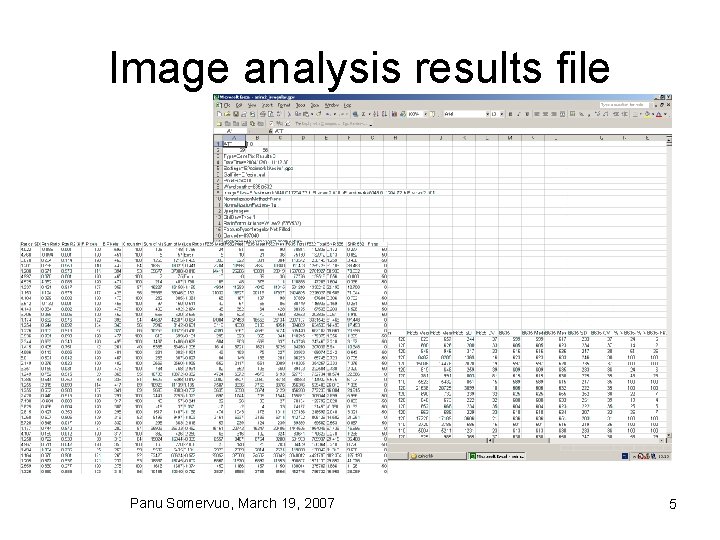
Image analysis results file Panu Somervuo, March 19, 2007 5

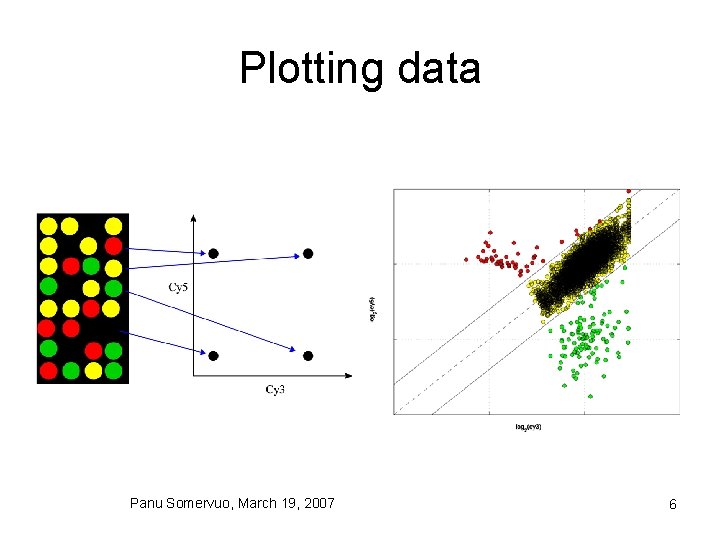
Plotting data Panu Somervuo, March 19, 2007 6

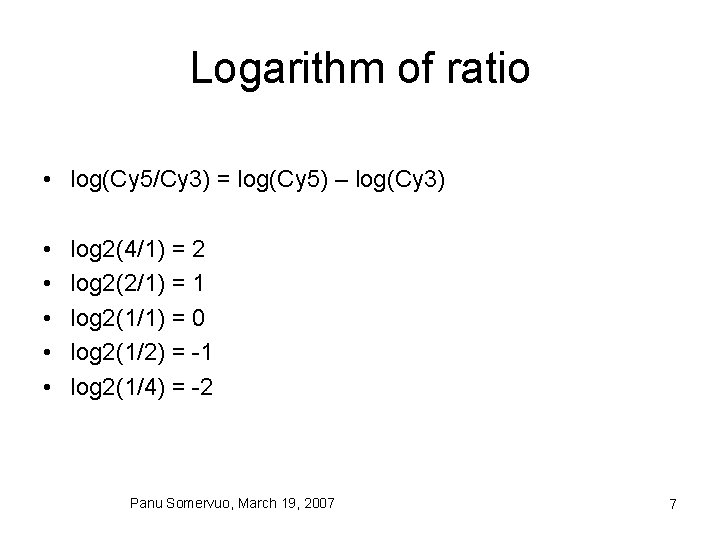
Logarithm of ratio • log(Cy 5/Cy 3) = log(Cy 5) – log(Cy 3) • • • log 2(4/1) = 2 log 2(2/1) = 1 log 2(1/1) = 0 log 2(1/2) = -1 log 2(1/4) = -2 Panu Somervuo, March 19, 2007 7

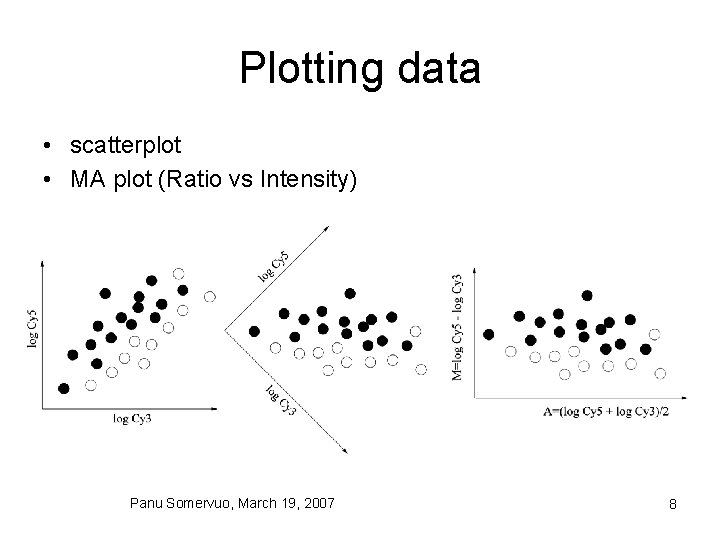
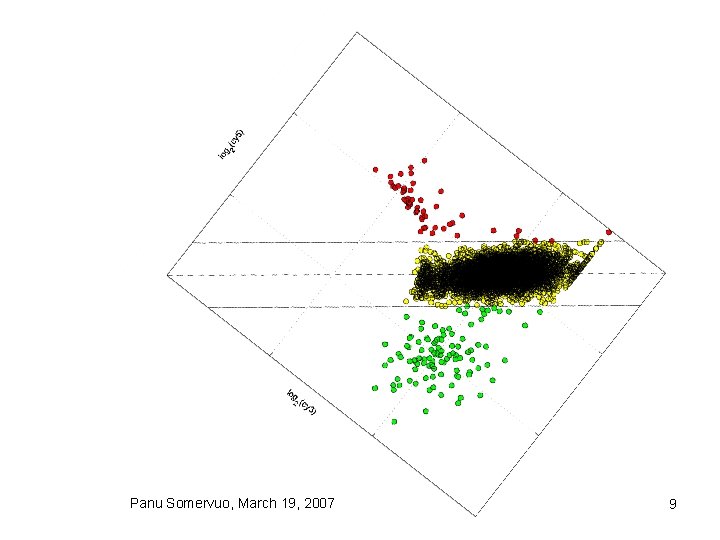
Plotting data • scatterplot • MA plot (Ratio vs Intensity) Panu Somervuo, March 19, 2007 8

Panu Somervuo, March 19, 2007 9

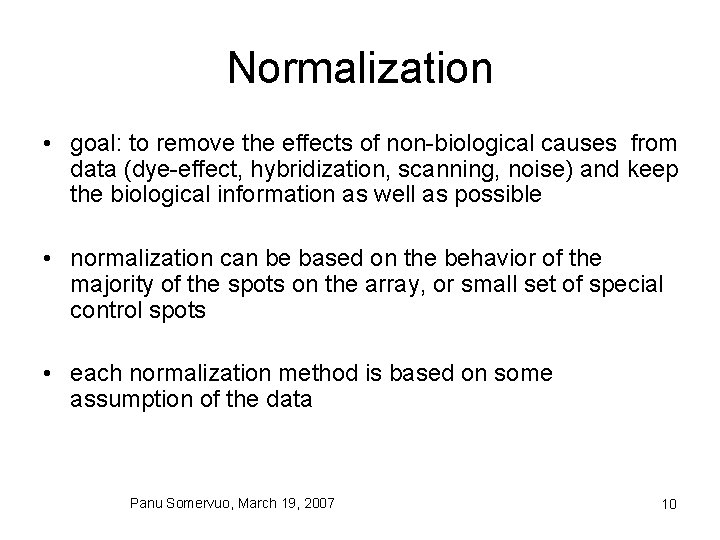
Normalization • goal: to remove the effects of non-biological causes from data (dye-effect, hybridization, scanning, noise) and keep the biological information as well as possible • normalization can be based on the behavior of the majority of the spots on the array, or small set of special control spots • each normalization method is based on some assumption of the data Panu Somervuo, March 19, 2007 10

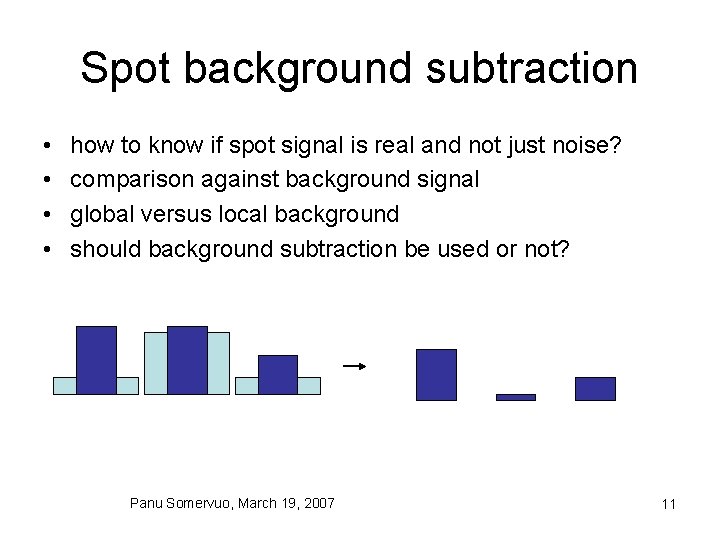
Spot background subtraction • • how to know if spot signal is real and not just noise? comparison against background signal global versus local background should background subtraction be used or not? Panu Somervuo, March 19, 2007 11

Normalization • can be applied to both single channel and ratio data • mean • variance Panu Somervuo, March 19, 2007 12

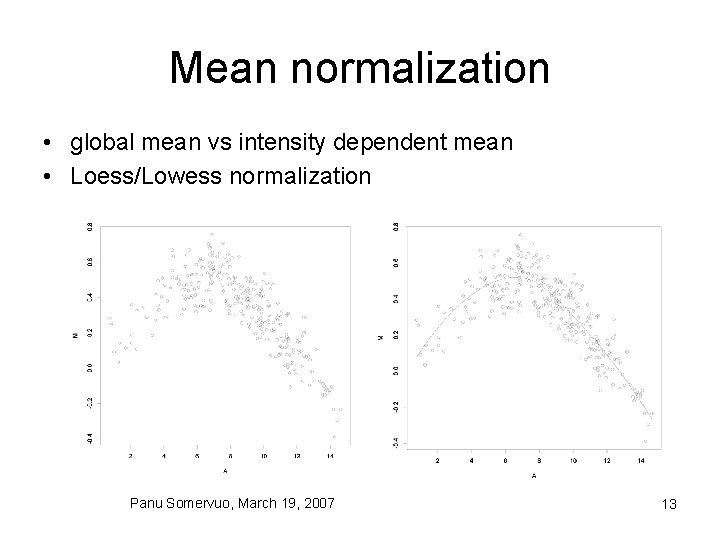
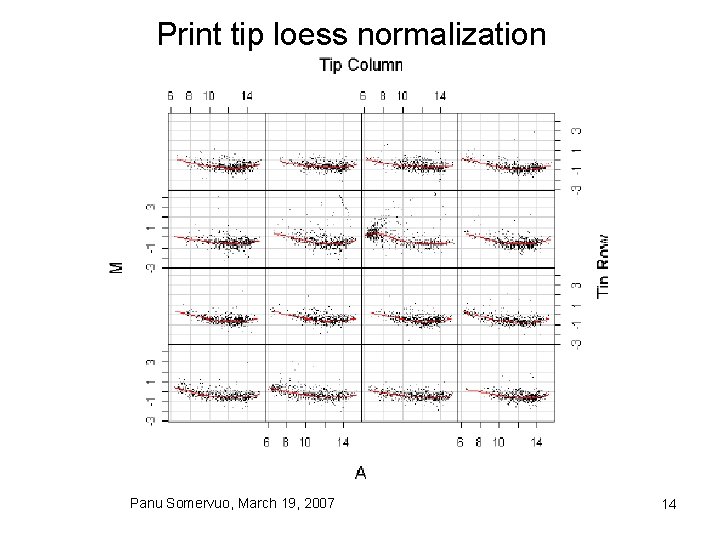
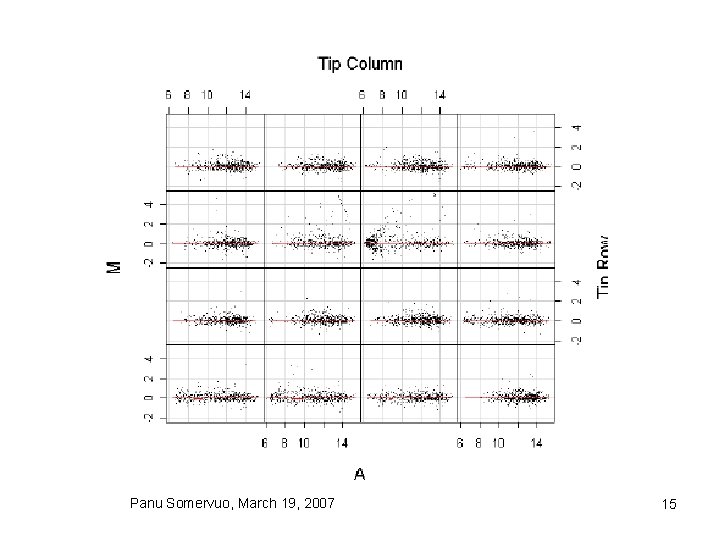
Mean normalization • global mean vs intensity dependent mean • Loess/Lowess normalization Panu Somervuo, March 19, 2007 13

Print tip loess normalization Panu Somervuo, March 19, 2007 14

Panu Somervuo, March 19, 2007 15

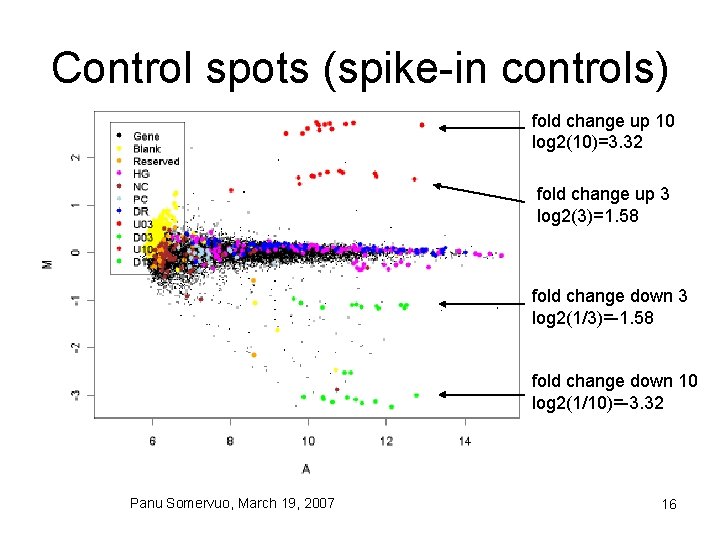
Control spots (spike-in controls) fold change up 10 log 2(10)=3. 32 fold change up 3 log 2(3)=1. 58 fold change down 3 log 2(1/3)=-1. 58 fold change down 10 log 2(1/10)=-3. 32 Panu Somervuo, March 19, 2007 16

What is the best normalization method? • each method is based on some assumption each method can fail • if utilizing the behavior of majority of the spots, array should represent all genes • if utilizing control spots, check if they are reliable • lots of methods have been introduced, lots of methods will be introduced… Panu Somervuo, March 19, 2007 17

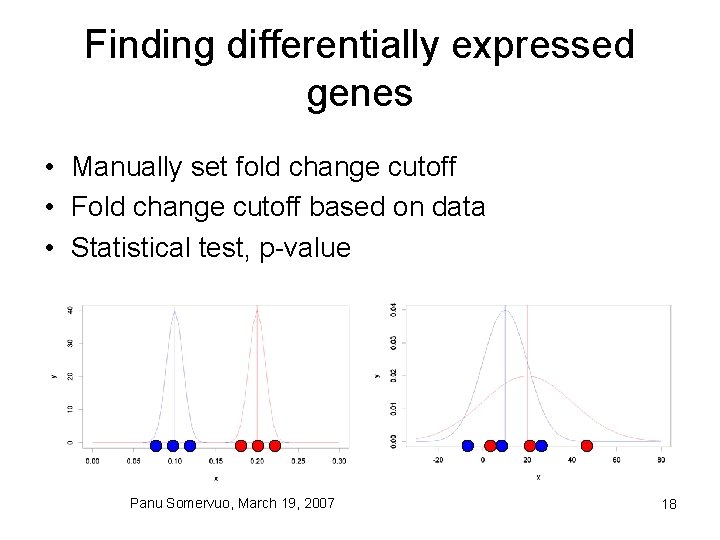
Finding differentially expressed genes • Manually set fold change cutoff • Fold change cutoff based on data • Statistical test, p-value Panu Somervuo, March 19, 2007 18

Limma package in R • analysis of microarray data – data import – data plotting – data normalization – statistical tests differentially expressed genes • online help and tutorial available > help(package=limma) > library(limma) > limma. Users. Guide() Panu Somervuo, March 19, 2007 19