Assessing Phylogenetic Hypotheses and Phylogenetic Data We use


















- Slides: 46

Assessing Phylogenetic Hypotheses and Phylogenetic Data • We use numerical phylogenetic methods because most data includes potentially misleading evidence of relationships • We should not be content with constructing phylogenetic hypotheses but should also assess what ‘confidence’ we can place in our hypotheses • This is not always simple! (but do not despair!)

Assessing Data Quality • We expect (or hope) our data will be well structured and contain strong phylogenetic signal • We can test this using randomisation tests of explicit null hypotheses • The behaviour or some measure of the quality of our real data is contrasted with that of comparable but phylogenetically uninformative data determined by randomisation of the data

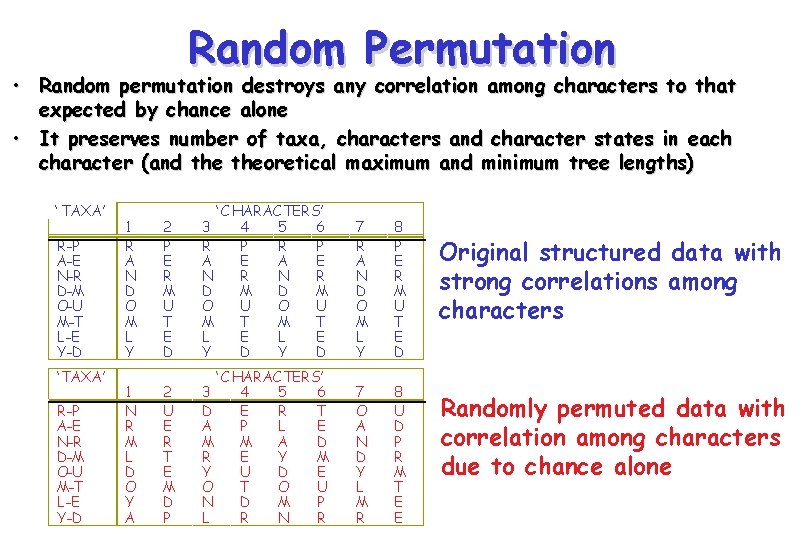
Random Permutation • Random permutation destroys any correlation among characters to that expected by chance alone • It preserves number of taxa, characters and character states in each character (and theoretical maximum and minimum tree lengths) ‘ TAXA’ R-P A-E N-R D-M O-U M-T L-E Y-D 1 R A N D O M L Y 1 N R M L D O Y A 2 P E R M U T E D 3 R A N D O M L Y ‘ CHARACTERS’ 4 5 6 P R P E A E R N R M D M U O U T M T E L E D Y D 7 R A N D O M L Y 8 P E R M U T E D 2 U E R T E M D P ‘ CHARACTERS’ 3 4 5 6 D E R T A P L E M M A D R E Y M Y U D E O T O U N D M P L R N R 7 O A N D Y L M R 8 U D P R M T E E Original structured data with strong correlations among characters Randomly permuted data with correlation among characters due to chance alone

Matrix Randomisation Tests • Compare some measure of data quality (hierarchical structure) for the real and many randomly permuted data sets • This allows us to define a test statistic for the null hypothesis that the real data are no better structured than randomly permuted and phylogenetically uninformative data • A permutation tail probability (PTP) is the proportion of data sets with as good or better measure of quality than the real data

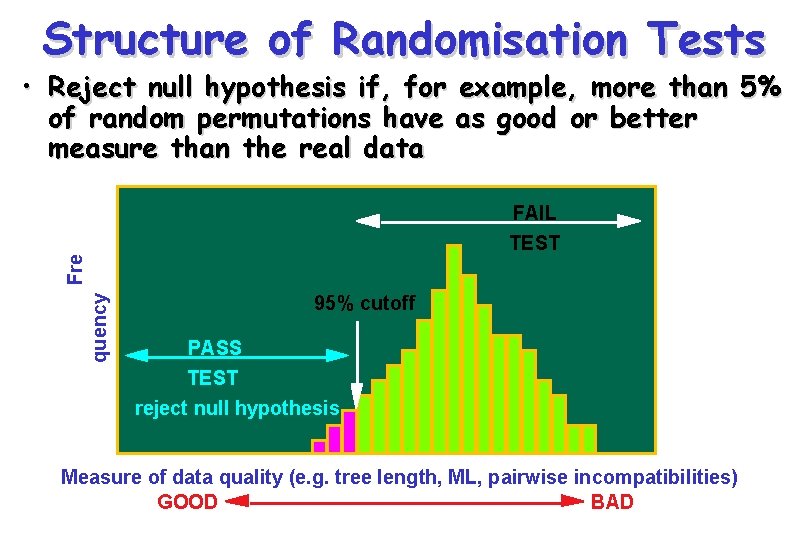
Structure of Randomisation Tests • Reject null hypothesis if, for example, more than 5% of random permutations have as good or better measure than the real data quency Fre FAIL TEST 95% cutoff PASS TEST reject null hypothesis Measure of data quality (e. g. tree length, ML, pairwise incompatibilities) GOOD BAD

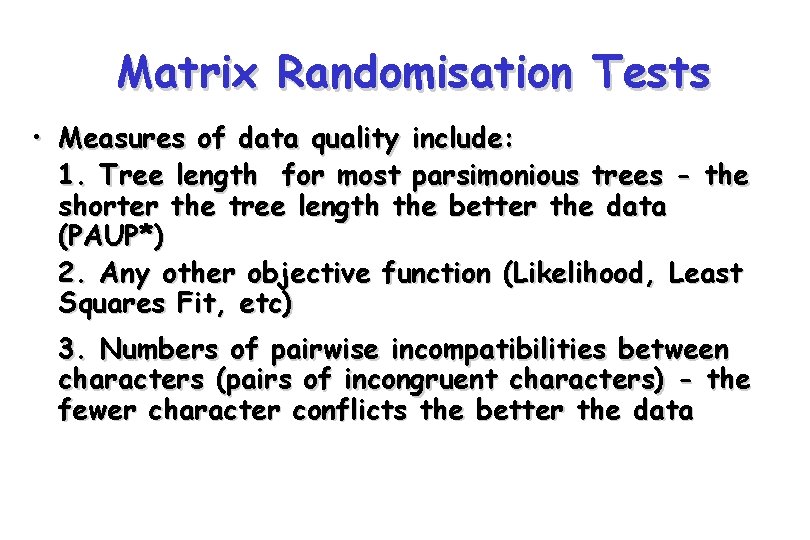
Matrix Randomisation Tests • Measures of data quality include: 1. Tree length for most parsimonious trees - the shorter the tree length the better the data (PAUP*) 2. Any other objective function (Likelihood, Least Squares Fit, etc) 3. Numbers of pairwise incompatibilities between characters (pairs of incongruent characters) - the fewer character conflicts the better the data

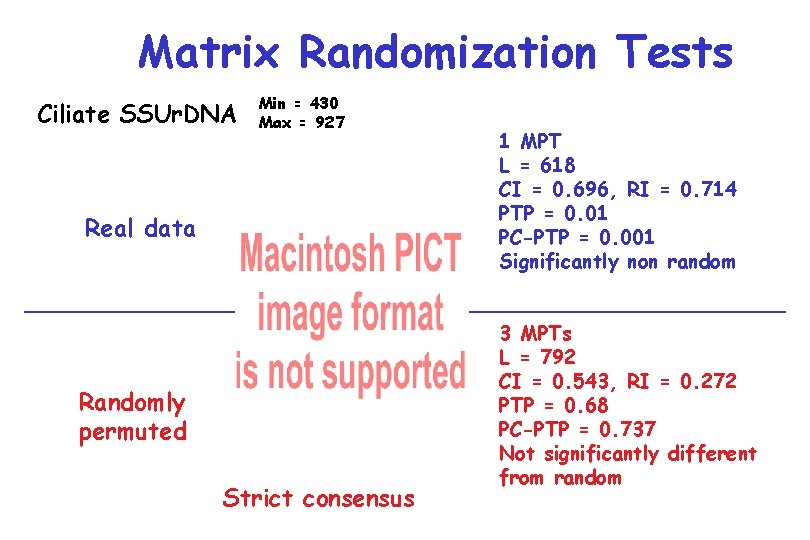
Matrix Randomization Tests Ciliate SSUr. DNA Min = 430 Max = 927 Real data Randomly permuted Strict consensus 1 MPT L = 618 CI = 0. 696, RI = 0. 714 PTP = 0. 01 PC-PTP = 0. 001 Significantly non random 3 MPTs L = 792 CI = 0. 543, RI = 0. 272 PTP = 0. 68 PC-PTP = 0. 737 Not significantly different from random

Matrix Randomisation Tests - use and limitations • Can detect very poor data - that provides no good basis for phylogenetic inferences (throw it away!) • However, only very little may be needed to reject the null hypothesis (passing test great data) • Doesn’t indicate location of this structure (more discerning tests are possible)

Assessing Phylogenetic Hypotheses - groups on trees • Several methods have been proposed that attach numerical values to internal branches in trees that are intended to provide some measure of the strength of support for those branches and the corresponding groups • These methods include: character resampling methods - the bootstrap and jackknife comparisons with suboptimal trees - decay analyses additional randomisation tests

Bootstrapping (non-parametric) • Bootstrapping is a modern statistical technique that uses computer intensive random resampling of data to determine sampling error or confidence intervals for some estimated parameter

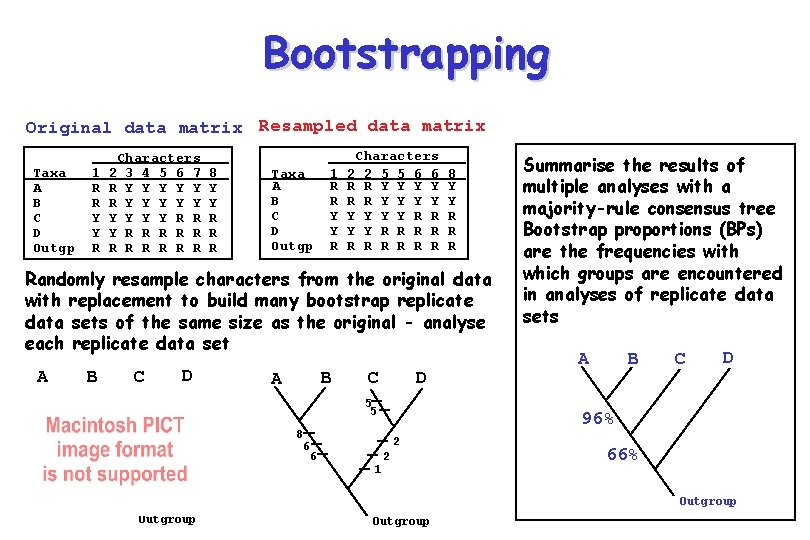
Bootstrapping • Characters are resampled with replacement to create many bootstrap replicate data sets • Each bootstrap replicate data set is analysed (e. g. with parsimony, distance, ML) • Agreement among the resulting trees is summarized with a majority-rule consensus tree • Frequency of occurrence of groups, bootstrap proportions (BPs), is a measure of support for those groups • Additional information is given in partition tables

Bootstrapping Original data matrix Resampled data matrix Taxa A B C D Outgp 1 R R Y Y R Characters 2 3 4 5 6 7 R Y Y Y Y R R R R R R 8 Y Y R R R Taxa A B C D Outgp 1 R R Y Y R Characters 2 2 5 5 6 6 R R Y Y Y R R Y Y R R R R R 8 Y Y R Randomly resample characters from the original data with replacement to build many bootstrap replicate data sets of the same size as the original - analyse each replicate data set A B C 1 2 8 7 6 D 1 2 5 4 3 B A C D 5 5 8 6 6 A B C D 96% 2 1 Summarise the results of multiple analyses with a majority-rule consensus tree Bootstrap proportions (BPs) are the frequencies with which groups are encountered in analyses of replicate data sets 2 66% Outgroup

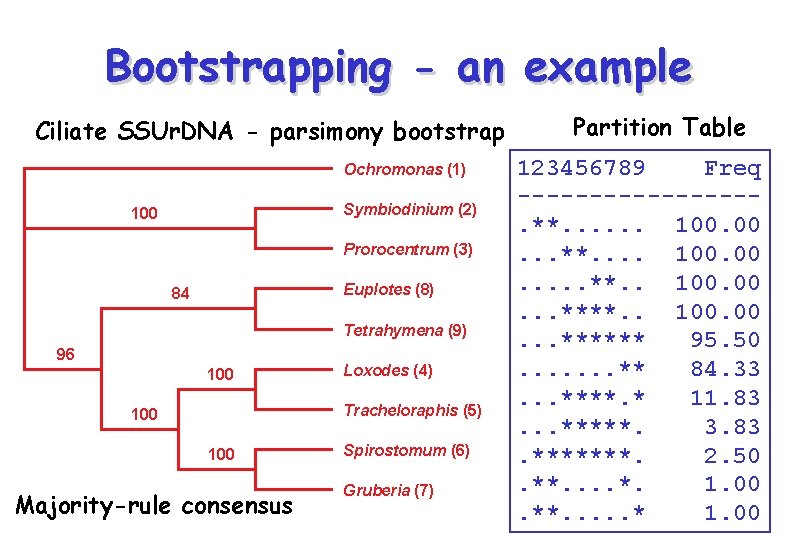
Bootstrapping - an example Ciliate SSUr. DNA - parsimony bootstrap Ochromonas (1) Symbiodinium (2) 100 Prorocentrum (3) Euplotes (8) 84 Tetrahymena (9) 96 100 Loxodes (4) Tracheloraphis (5) 100 Majority-rule consensus Spirostomum (6) Gruberia (7) Partition Table 123456789 Freq --------. **. . . 100. . . **. . 100. 00. . . ****** 95. 50. . . . ** 84. 33. . . ****. * 11. 83. . . *****. 3. 83. *******. 2. 50. **. . *. 1. 00. **. . . * 1. 00

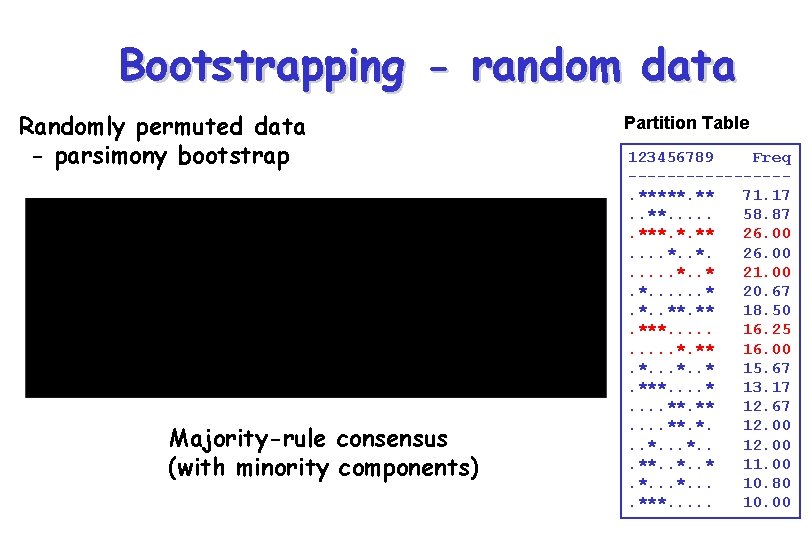
Bootstrapping - random data Randomly permuted data - parsimony bootstrap Majority-rule consensus (with minority components) Partition Table 123456789 Freq --------. *****. ** 71. 17. . **. . . 58. 87. ***. *. ** 26. 00. . *. 26. 00. . . * 21. 00. *. . . * 20. 67. *. . ** 18. 50. ***. . . 16. 25. . . *. ** 16. 00. *. . * 15. 67. ***. . * 13. 17. . **. ** 12. 67. . **. *. 12. 00. . *. . 12. 00. **. . * 11. 00. *. . . 10. 80. ***. . . 10. 00

Bootstrap - interpretation • Bootstrapping was introduced as a way of establishing confidence intervals for phylogenies • This interpretation of bootstrap proportions (BPs) depends on assuming that the original data is a random (fair) sample from independent and identically distributed data • However, several things complicate this interpretation - Perhaps the assumptions are unreasonable - making any statistical interpretation of BPs invalid - Some theoretical work indicates that BPs are very conservative, and may underestimate confidence intervals - problem increases with numbers of taxa - BPs can be high for incongruent relationships in separate analyses - and can therefore be misleading (misleading data -> misleading BPs) - with parsimony it may be highly affected by inclusion or exclusion of only a few characters

Bootstrap - interpretation • Bootstrapping is a very valuable and widely used technique it (or some suitable) alternative is demanded by some journals, but it may require a pragmatic interpretation: • BPs depend on two aspects of the support for a group - the numbers of characters supporting a group and the level of support for incongruent groups • BPs thus provides an index of the relative support for groups provided by a set of data under whatever interpretation of the data (method of analysis) is used

Bootstrap - interpretation • High BPs (e. g. > 85%) is indicative of strong ‘signal’ in the data • Provided we have no evidence of strong misleading signal (e. g. base composition biases, great differences in branch lengths) high BPs are likely to reflect strong phylogenetic signal • Low BPs need not mean the relationship is false, only that it is poorly supported • Bootstrapping can be viewed as a way of exploring the robustness of phylogenetic inferences to perturbations in the balance of supporting and conflicting evidence for groups

Jackknifing • Jackknifing is very similar to bootstrapping and differs only in the character resampling strategy • Some proportion of characters (e. g. 50%) are randomly selected and deleted • Replicate data sets are analysed and the results summarised with a majority-rule consensus tree • Jackknifing and bootstrapping tend to produce broadly similar results and have similar interpretations

Decay analysis • In parsimony analysis, a way to assess support for a group is to see if the group occurs in slightly less parsimonious trees also • The length difference between the shortest trees including the group and the shortest trees that exclude the group (the extra steps required to overturn a group) is the decay index or Bremer support • Can be extended to any optimality criterion and to other relationships

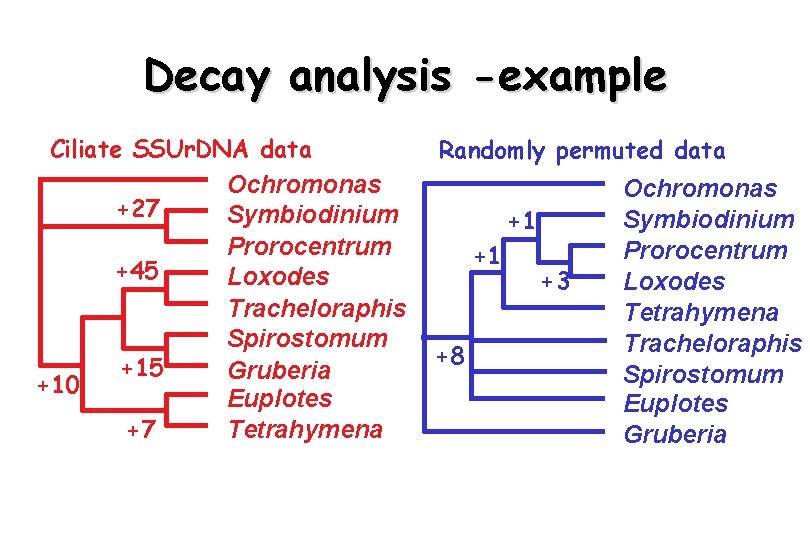
Decay analysis -example Ciliate SSUr. DNA data Ochromonas +27 Symbiodinium Prorocentrum +45 Loxodes Tracheloraphis Spirostomum +15 Gruberia +10 Euplotes +7 Tetrahymena Randomly permuted data +1 +1 +8 +3 Ochromonas Symbiodinium Prorocentrum Loxodes Tetrahymena Tracheloraphis Spirostomum Euplotes Gruberia

Decay indices - interpretation • Generally, the higher the decay index the better the relative support for a group • Like BPs, decay indices may be misleading if the data is misleading • Unlike BPs decay indices are not scaled (0 -100) and it is less clear what is an acceptable decay index • Magnitude of decay indices and BPs generally correlated (i. e. they tend to agree) • Only groups found in all most parsimonious trees have decay indices > zero

Types of Cladistic Relationships 5 -Taxon Tree

Extending Support Measures • The same measures (BP, JP & DI) that are used for clades/splits can also be determined for triplets and quartets • This provides a lot more information because there are more triplets/quartets than there are clades • Furthermore. .

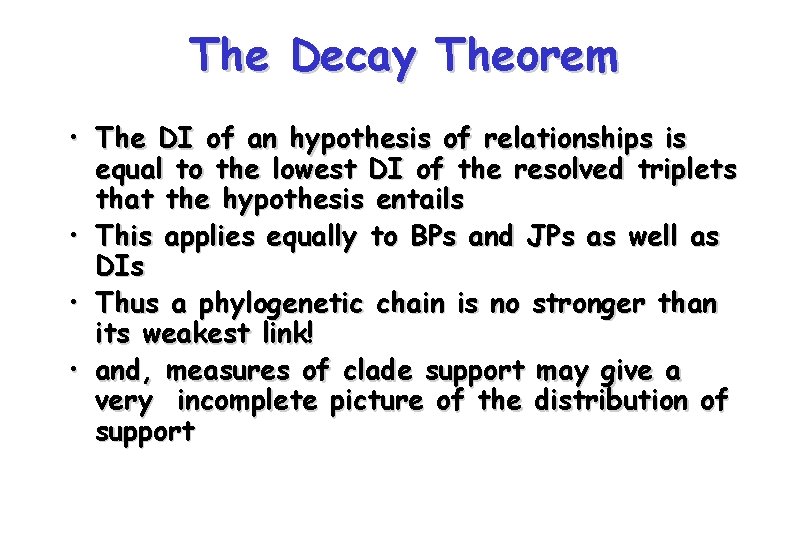
The Decay Theorem • The DI of an hypothesis of relationships is equal to the lowest DI of the resolved triplets that the hypothesis entails • This applies equally to BPs and JPs as well as DIs • Thus a phylogenetic chain is no stronger than its weakest link! • and, measures of clade support may give a very incomplete picture of the distribution of support

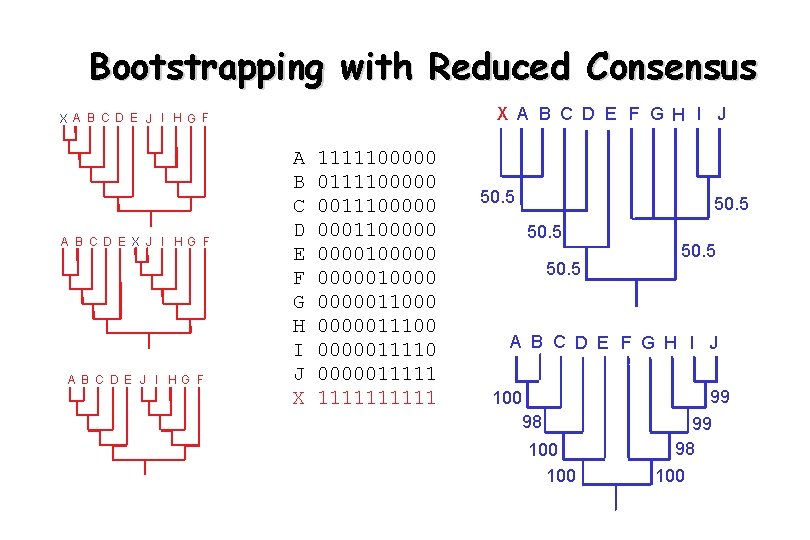
Bootstrapping with Reduced Consensus X A B C D E F G H I J X A B C D E J I HG F A B C D E X J I HG F A B C D E F G H I J X 1111100000 001110000010000011000 0000011110 0000011111 50. 5 A B C D E F G H I J 100 98 100 99 99 98 100


Leaf Stability • Leaf stability is the average of supports of the triplets/quartets containing the leaf

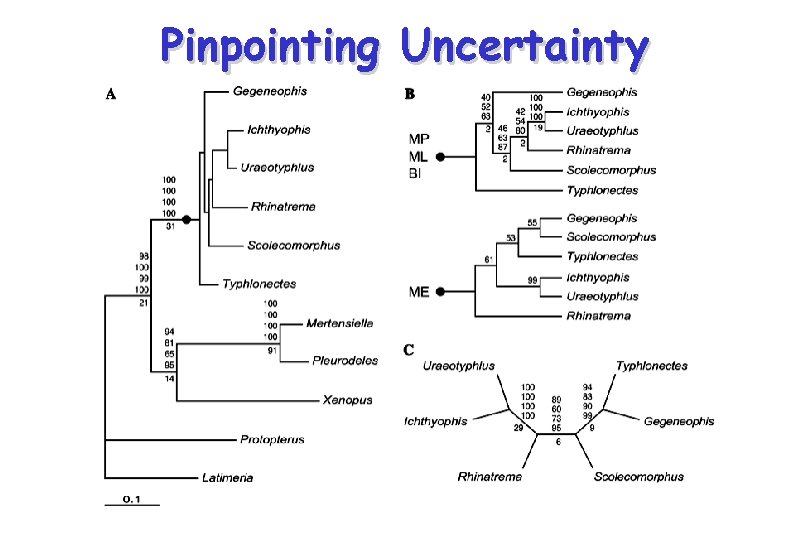
Pinpointing Uncertainty

Comparing competing hypotheses - tests of trees phylogenetic two (or more) • Particularly useful techniques are those designed to allow evaluation of alternative phylogenetic hypotheses • Several such tests allow us to determine if one tree is statistically significantly worse than another: Winning sites, Templeton, Kishino-Hasegawa, parametric bootstrapping (SOWH) Shimodaira-Hasegawa, Approximately Unbiased

Tests of two trees • Tests are of the null hypothesis that the differences between two trees (A and B) are no greater than expected from sampling error • The simplest ‘wining sites’ test sums the number of sites supporting tree A over tree B and vice versa (those having fewer steps on, and better fit to, one of the trees) • Under the null hypothesis characters are equally likely to support tree A or tree B and a binomial distribution gives the probability of the observed difference in numbers of winning sites

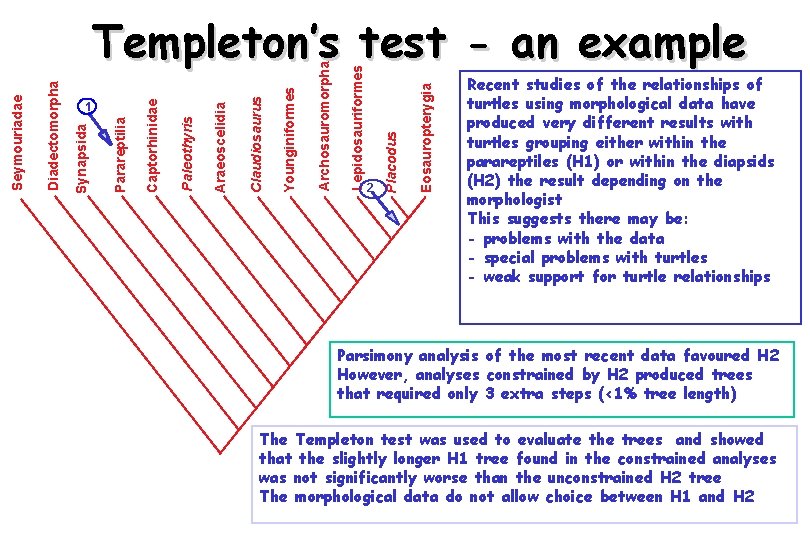
The Templeton test • Templeton’s test is a non-parametric Wilcoxon signed ranks test of the differences in fits of characters to two trees • It is like the ‘winning sites’ test but also takes into account the magnitudes of differences in the support of characters for the two trees

Eosauropterygia 2 Placodus Lepidosauriformes Archosauromorpha Younginiformes Claudiosaurus Araeoscelidia Paleothyris Captorhinidae Parareptilia 1 Synapsida Diadectomorpha Seymouriadae Templeton’s test - an example Recent studies of the relationships of turtles using morphological data have produced very different results with turtles grouping either within the parareptiles (H 1) or within the diapsids (H 2) the result depending on the morphologist This suggests there may be: - problems with the data - special problems with turtles - weak support for turtle relationships Parsimony analysis of the most recent data favoured H 2 However, analyses constrained by H 2 produced trees that required only 3 extra steps (<1% tree length) The Templeton test was used to evaluate the trees and showed that the slightly longer H 1 tree found in the constrained analyses was not significantly worse than the unconstrained H 2 tree The morphological data do not allow choice between H 1 and H 2

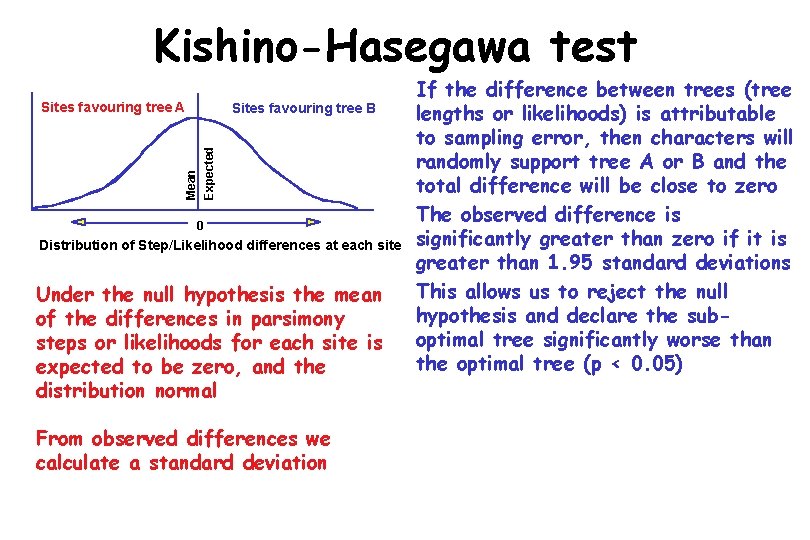
Kishino-Hasegawa test • The Kishino-Hasegawa test is similar in using differences in the support provided by individual sites for two trees to determine if the overall differences between the trees are significantly greater than expected from random sampling error • It is a parametric test that depends on assumptions that the characters are independent and identically distributed (the same assumptions underlying the statistical interpretation of bootstrapping) • It can be used with parsimony and maximum likelihood - implemented in PHYLIP and PAUP*

Kishino-Hasegawa test Sites favouring tree A Mean Expected Sites favouring tree B 0 Distribution of Step/Likelihood differences at each site Under the null hypothesis the mean of the differences in parsimony steps or likelihoods for each site is expected to be zero, and the distribution normal From observed differences we calculate a standard deviation If the difference between trees (tree lengths or likelihoods) is attributable to sampling error, then characters will randomly support tree A or B and the total difference will be close to zero The observed difference is significantly greater than zero if it is greater than 1. 95 standard deviations This allows us to reject the null hypothesis and declare the suboptimal tree significantly worse than the optimal tree (p < 0. 05)

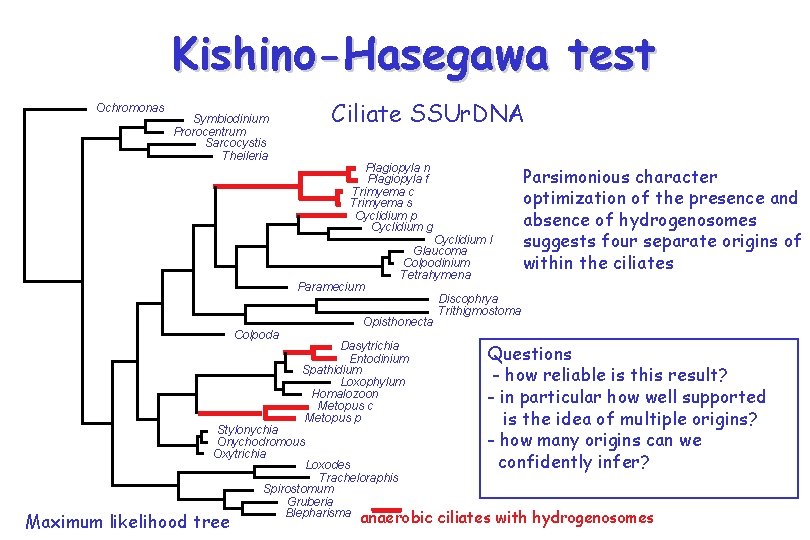
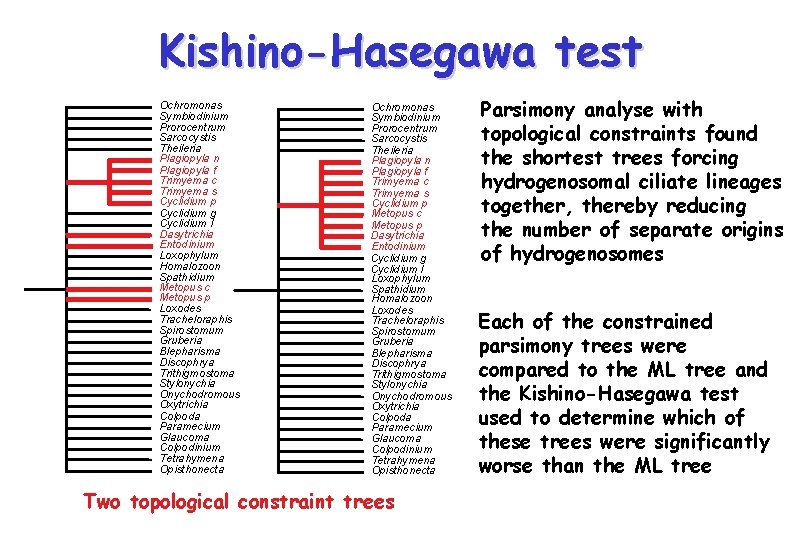
Kishino-Hasegawa test Ochromonas Symbiodinium Prorocentrum Sarcocystis Theileria Ciliate SSUr. DNA Plagiopyla n Plagiopyla f Trimyema c Trimyema s Cyclidium p Cyclidium g Paramecium Colpoda Opisthonecta Dasytrichia Entodinium Spathidium Loxophylum Homalozoon Metopus c Metopus p Stylonychia Onychodromous Oxytrichia Loxodes Tracheloraphis Spirostomum Gruberia Blepharisma Maximum likelihood tree Cyclidium l Glaucoma Colpodinium Tetrahymena Parsimonious character optimization of the presence and absence of hydrogenosomes suggests four separate origins of within the ciliates Discophrya Trithigmostoma Questions - how reliable is this result? - in particular how well supported is the idea of multiple origins? - how many origins can we confidently infer? anaerobic ciliates with hydrogenosomes

Kishino-Hasegawa test Ochromonas Symbiodinium Prorocentrum Sarcocystis Theileria Plagiopyla n Plagiopyla f Trimyema c Trimyema s Cyclidium p Cyclidium g Cyclidium l Dasytrichia Entodinium Loxophylum Homalozoon Spathidium Metopus c Metopus p Loxodes Tracheloraphis Spirostomum Gruberia Blepharisma Discophrya Trithigmostoma Stylonychia Onychodromous Oxytrichia Colpoda Paramecium Glaucoma Colpodinium Tetrahymena Opisthonecta Ochromonas Symbiodinium Prorocentrum Sarcocystis Theileria Plagiopyla n Plagiopyla f Trimyema c Trimyema s Cyclidium p Metopus c Metopus p Dasytrichia Entodinium Cyclidium g Cyclidium l Loxophylum Spathidium Homalozoon Loxodes Tracheloraphis Spirostomum Gruberia Blepharisma Discophrya Trithigmostoma Stylonychia Onychodromous Oxytrichia Colpoda Paramecium Glaucoma Colpodinium Tetrahymena Opisthonecta Two topological constraint trees Parsimony analyse with topological constraints found the shortest trees forcing hydrogenosomal ciliate lineages together, thereby reducing the number of separate origins of hydrogenosomes Each of the constrained parsimony trees were compared to the ML tree and the Kishino-Hasegawa test used to determine which of these trees were significantly worse than the ML tree

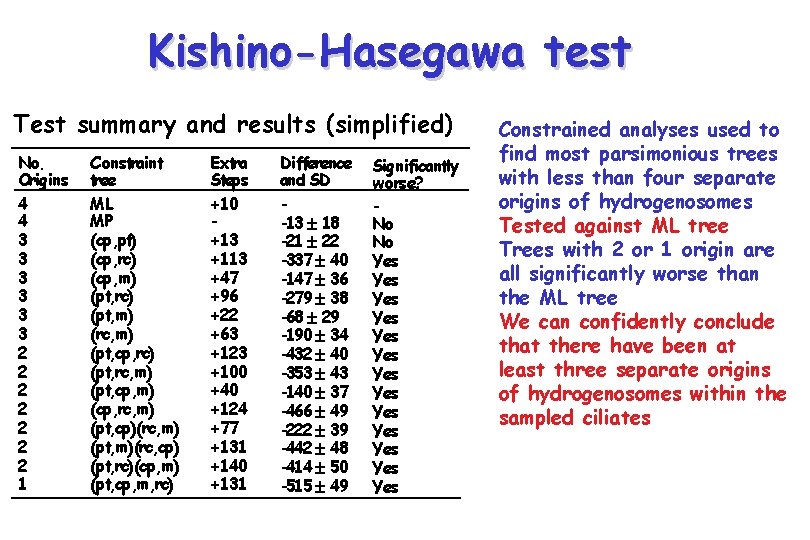
Kishino-Hasegawa test Test summary and results (simplified) No. Origins 4 4 3 3 3 2 2 2 2 1 Constraint tree ML MP (cp, pt) (cp, rc) (cp, m) (pt, rc) (pt, m) (rc, m) (pt, cp, rc) (pt, rc, m) (pt, cp, m) (cp, rc, m) (pt, cp)(rc, m) (pt, m)(rc, cp) (pt, rc)(cp, m) (pt, cp, m, rc) Extra Steps +10 +13 +113 +47 +96 +22 +63 +123 +100 +40 +124 +77 +131 +140 +131 Difference and SD -13 18 -21 22 -337 40 -147 36 -279 38 -68 29 -190 34 -432 40 -353 43 -140 37 -466 49 -222 39 -442 48 -414 50 -515 49 Significantly worse? No No Yes Yes Yes Yes Constrained analyses used to find most parsimonious trees with less than four separate origins of hydrogenosomes Tested against ML tree Trees with 2 or 1 origin are all significantly worse than the ML tree We can confidently conclude that there have been at least three separate origins of hydrogenosomes within the sampled ciliates

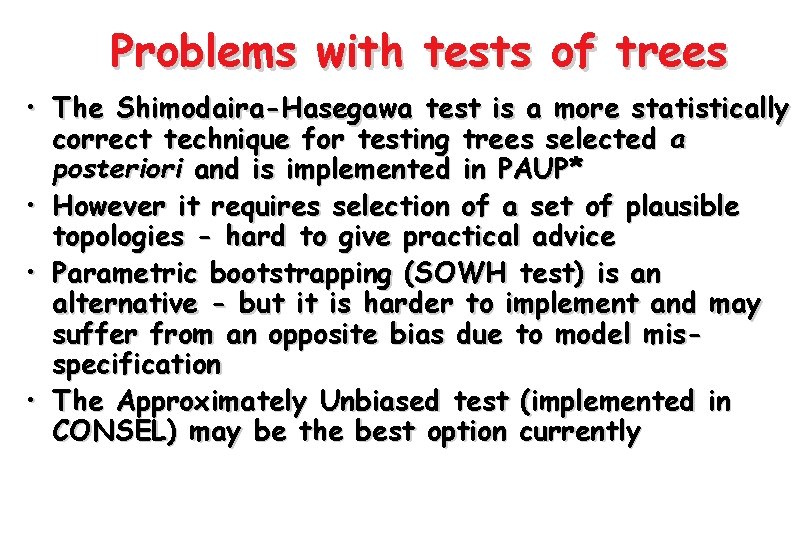
Problems with tests of trees • To be statistically valid, the Kishino-Hasegawa test should be of trees that are selected a priori • However, most applications have used trees selected a posteriori on the basis of the phylogenetic analysis • Where we test the ‘best’ tree against some other tree the KH test will be biased towards rejection of the null hypothesis • Only if null hypothesis is not rejected will result be safe from some unknown level of bias

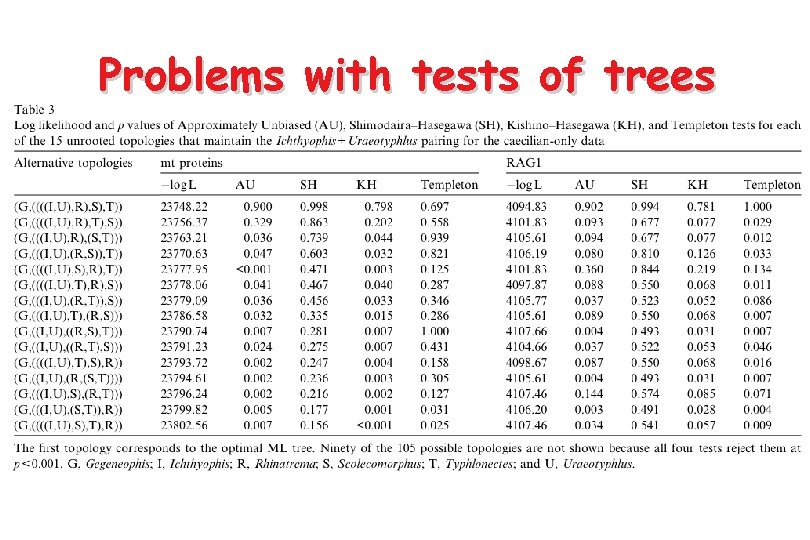
Problems with tests of trees • The Shimodaira-Hasegawa test is a more statistically correct technique for testing trees selected a posteriori and is implemented in PAUP* • However it requires selection of a set of plausible topologies - hard to give practical advice • Parametric bootstrapping (SOWH test) is an alternative - but it is harder to implement and may suffer from an opposite bias due to model misspecification • The Approximately Unbiased test (implemented in CONSEL) may be the best option currently

Problems with tests of trees

Taxonomic Congruence • Trees inferred from different data sets (different genes, morphology) should agree if they are accurate • Congruence between trees is best explained by their accuracy • Congruence can be investigated using consensus (and supertree) methods • Incongruence requires further work to explain or resolve disagreements

Reliability of Phylogenetic Methods • Phylogenetic methods (e. g. parsimony, distance, ML) can also be evaluated in terms of their general performance, particularly their: consistency - approach the truth with more data efficiency - how quickly (how much data) robustness - sensitivity to violations of assumptions • Studies of these properties can be analytical or by simulation

Reliability of Phylogenetic Methods • There have been many arguments that ML methods are best because they have desirable statistical properties, such as consistency • However, ML does not always have these properties – if the model is wrong/inadequate (fortunately this is testable to some extent) – properties not yet demonstrated for complex inference problems such as phylogenetic trees

Reliability of Phylogenetic Methods • “Simulations show that ML methods generally outperform distance and parsimony methods over a broad range of realistic conditions” Whelan et al. 2001 Trends in Genetics 17: 262 -272 • But… • Most simulations cover a narrow range of very (unrealistically) simple conditions – few taxa (typically just four!) – few parameters (standard models - JC, K 2 P etc)

Reliability of Phylogenetic Methods • Simulations with four taxa have shown: - Model based methods - distance and maximum likelihood perform well when the model is accurate (not surprising!) - Violations of assumptions can lead to inconsistency for all methods (a Felsenstein zone) when branch lengths or rates are highly unequal - Maximum likelihood methods are quite robust to violations of model assumptions - Weighting can improve the performance of parsimony (reduce the size of the Felsenstein zone)

Reliability of Phylogenetic Methods • However: - Generalising from four taxon simulations may be dangerous as conclusions may not hold for more complex cases - A few large scale simulations (many taxa) have suggested that parsimony can be very accurate and efficient - Most methods are accurate in correctly recovering known phylogenies produced in laboratory studies • More realistic simulations are needed if they are to help in choosing/understanding methods • You can try your own…