Phylogenetic Tree 2272021 1 Phylogenetic Tree What it























- Slides: 56

Phylogenetic Tree 2/27/2021 1

Phylogenetic Tree: What it is n n n 2/27/2021 Drawing evolutionary tree from characteristics of organisms or some measured distances between them Represented as a tree where nodes are the organisms/objects and arcs are the proximity between the respective nodes Based on how close the organisms are 2

Phylogenetic Tree: Motivation n n Pure curiosity: biological science One species can be studied for a related one: u u n n n 2/27/2021 Drug test on monkeys for human Rare species can be spared in a study Drug design on evolution of microorganism: aids/flu vaccine/drug design depends on how do they evolve Tracking pathogen sources Genesis, archeology, , , 3

Phylogenetic Tree: topology n n n 2/27/2021 Evolutionary distance is not same as elapsed time: former is a crude approximation of the latter (if distance can be calculated at all) Leaves are objects, internal nodes may or may not be objects (may represent hypothetical ancestors) Mostly binary trees, sometimes not 4

Phylogenetic Tree: source data types n Discrete characters: u u u n Numerical distance matrix: u n 2/27/2021 does it have long beaks? Could be Boolean or multi-valued Provided in matrix form (objects X characters) Symmetric pairwise distances measured by some means, e. g. , by aligning sequences Continuous character: character value is in numerical domain 5

Characters for phylogeny n n 2/27/2021 Characters should be relevant in the context of phylogeny: depends on the user scientist Characters should be independent: inherited without interference between the characters (eye color and hair color may not be a good combination in character set) All characters must evolve from the same ancestor: we presume that (1) it is tree, (2) it is a connected tree Closest objects are called “homologous”: max possible characters have same 6 values or related values

Phylogeny using character state matrix n n 2/27/2021 A “state” is a tuple with values for each character (value could be “unassigned”) Internal node may be a state without any object assigned on it Leaves are where the states correspond to objects with the respective assigned characters P 178: a source character state matrix 7

Phylogeny using character state matrix: Problems n 2/27/2021 Convergence evolution: two nonhomologous objects (most characters does not match, loosely speaking) happen to have same value on a character (needs a cycle in the graph) 8

Phylogeny using character state matrix: Problems n n 2/27/2021 In one case evolution suggests character value of c evolves from “long” to “short, ” in another case the reverse: confusion over the direction of evolution Again, the tree property would be violated to accommodate this 9

Character domain types Domain of character c could be: red < - > blue < - > yellow < - > green n n 2/27/2021 C cannot evolve from blue to green without taking value yellow first C is “ordered” C can be directed and ordered, instead of undirected as above 10

Perfect phylogeny n n 2/27/2021 Noise-free input Each edge in phylogeny is a transition of the respective character’s value All nodes with the same value for a character must form a subtree (with the transition at its root) Such a tree is “perfect phylogeny” 11

Perfect phylogeny problem n n 2/27/2021 Given a character state matrix does there exist a perfect phylogeny over it P 178 table does not have a perfect phylogeny (presume transitions always 0 -> 1). Why? P 180: table and its perfect phylogeny What do you do when you do not have perfect phylogeny? Presume data is noisy and minimize errors in drawing perfect phylogeny 12

Perfect phylogeny problem n n You can always try all possible trees over the objects and check whether each tree is perfect phylogeny or not The total number of such trees is π 2/27/2021 i=3 n (2 i-5): Exponential 13

Perfect phylogeny problem: to check existence (Boolean matrix) 2/27/2021 14

Perfect phylogeny problem: to check existence (Boolean matrix) n n Organize char state matrix colum-wise: for each col i set of objects is Oi Every pair of Oi and Ok should be: u u u n n 2/27/2021 either Oi Ok or Oi Ok = null Either one belongs to the other one or they do not overlap at all If they overlap, no perfect phylogeny exist 15

Perfect phylogeny problem: to check existence (Boolean matrix) n n n 2/27/2021 In contrary, suppose Oi and Ok overlaps and a perfect phylogeny exists say, i is the edge between (u, v): v and subtree has i=1, but all other nodes have i=0. Suppose, three objects a, b, and c such that, a, b Oi, but c is not: a, b in subtree of v and c is not there But, suppose b, c Ok, and a is not: b, c must belong to some other subtree separated by edge k Contradiction 16

Perfect phylogeny problem: to check existence (Boolean matrix) n When no overlap exists: u u n n 2/27/2021 Contained sets go within same subtree, if Oi Ok, then i-subtree is subtree of ksubtree Disjoint sets are separate subtrees Proves if and only if of the condition for perfect phylogeny Algorithm for checking: Pairwise checking of object set may take O(m^2) for m characters, but set overlap may check even more time 17

Perfect phylogeny problem: Algorithm (Boolean matrix) n n 2/27/2021 Sort the columns by number of 1’s (descending) Scan each row to find which col number has the rightmost 1 for that box Scan each column: every box should agree Complexity O(mn) count, O(m log m) sort, O(mn) index matrix creation, O(mn) checking over index matrix: total O(mn) presuming n > log m 18

Perfect phylogeny problem: Algorithm (Boolean matrix) n n 2/27/2021 Exercise: try the algorithm for tables 6. 1 p 178 and 6. 2 p 180 Construction Algorithm: (1) sort characters/col increasing order, (2) each object – (3) each character – (4) if edge for char exists put obj on the end, (5) else create an edge and put object at the end, (6: cosmetic step) if more objects in a leaf node create edges for each object O(nm) Exc. Try it on table 6. 2 p 180 19

Perfect phylogeny problem: Algorithm (non-Boolean matrix, but…) n If two states per character but the order of transition not known, then presume an order: u n 2/27/2021 majority state 0, minority 1 (more ancestors are available) Same Lemma must be applied after this presumption: no overlapping set of objects 20

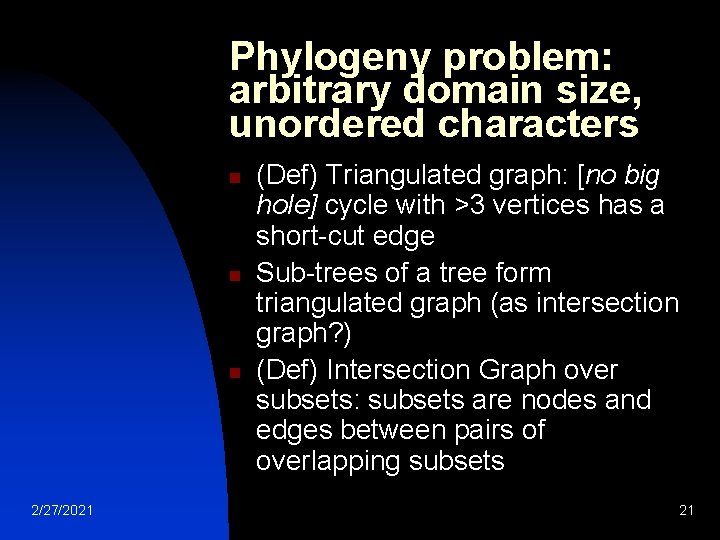
Phylogeny problem: arbitrary domain size, unordered characters n n n 2/27/2021 (Def) Triangulated graph: [no big hole] cycle with >3 vertices has a short-cut edge Sub-trees of a tree form triangulated graph (as intersection graph? ) (Def) Intersection Graph over subsets: subsets are nodes and edges between pairs of overlapping subsets 21

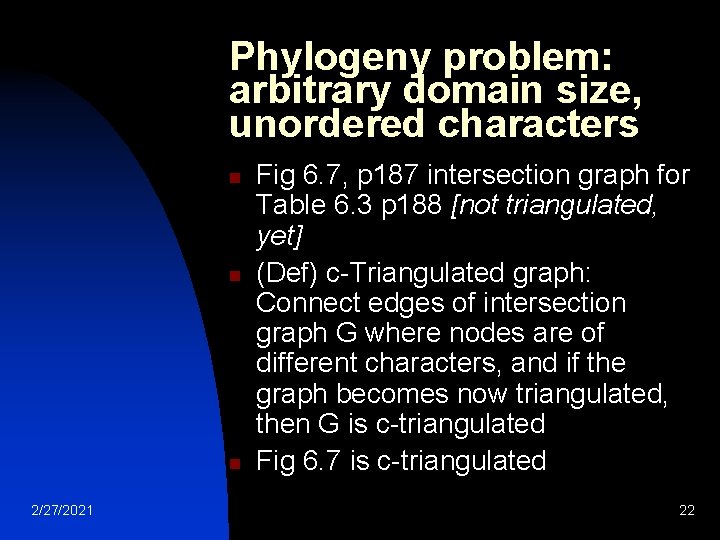
Phylogeny problem: arbitrary domain size, unordered characters n n n 2/27/2021 Fig 6. 7, p 187 intersection graph for Table 6. 3 p 188 [not triangulated, yet] (Def) c-Triangulated graph: Connect edges of intersection graph G where nodes are of different characters, and if the graph becomes now triangulated, then G is c-triangulated Fig 6. 7 is c-triangulated 22

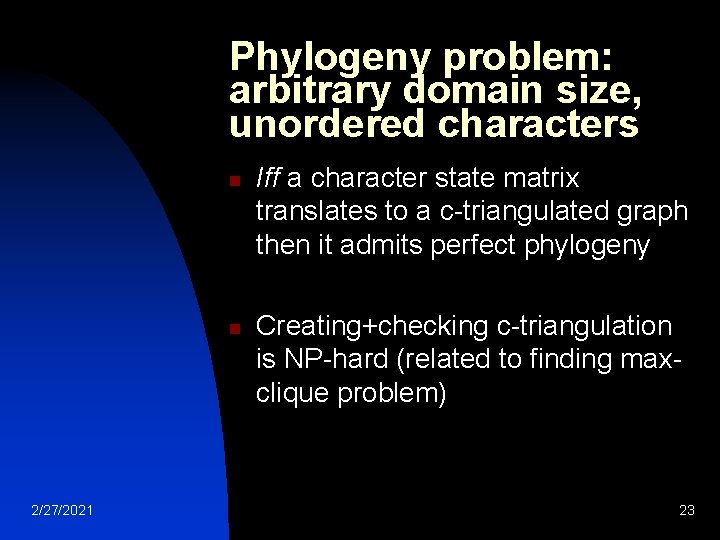
Phylogeny problem: arbitrary domain size, unordered characters n n 2/27/2021 Iff a character state matrix translates to a c-triangulated graph then it admits perfect phylogeny Creating+checking c-triangulation is NP-hard (related to finding maxclique problem) 23

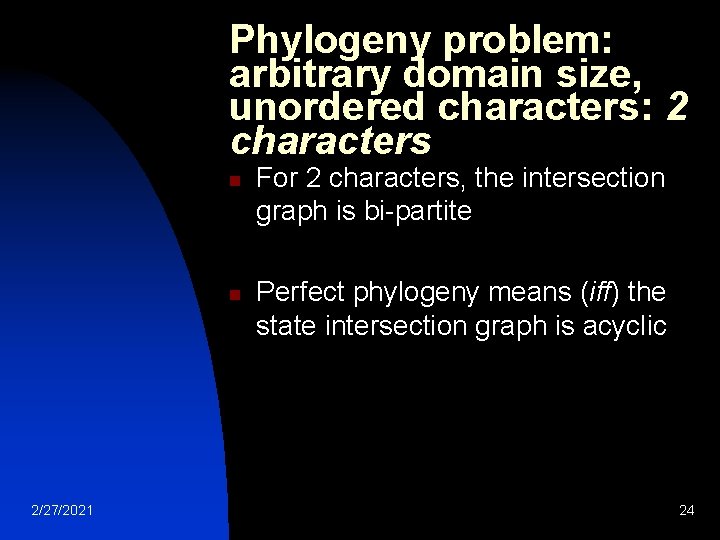
Phylogeny problem: arbitrary domain size, unordered characters: 2 characters n n 2/27/2021 For 2 characters, the intersection graph is bi-partite Perfect phylogeny means (iff) the state intersection graph is acyclic 24

Phylogeny construction: arbitrary domain size, unordered characters: 2 characters n Algorithm: u u u n 2/27/2021 (1) Construct intersection graph (2) make nodes for edges (intersection of the objects in old nodes now goes to the new nodes) (3) connect new nodes if they have overlapping objects (4) spanning tree of the graph is phylogeny (5: cosmetic step) objects huddled on a node should be put on separate leaves Try on Table 6. 4 p 190, and check against Fig 6. 8 p 189 25

When Perfect Phylogeny does not exist n n n 2/27/2021 Eliminate problematic characters: which ones, an optimization problem – min number of characters: Compatibility criterion Minimize convergence (character goes back to its previous value): Parsimony criterion Both NP-complete problems 26

When Perfect Phylogeny does not exist: Parsimony n n 2/27/2021 Compatibility problem: Does there exist a subset of characters such that Lemma 6. 1 (non-overlapping set of objects) is valid (or Perfect Phylogeny exists)? Equivalent to K-clique problem: does there exist a connectedsubgraph with K or more nodes? 27

When Perfect Phylogeny does not exist: Parsimony n n n 2/27/2021 Poly-transformation from Clique to compatibility problem: nodes to character, 3 objects for each edge with specific character values Every pair of NP-complete problems have two way poly-trans Compatibility can also be poly-trans to Clique: characters to nodes, nonoverlapping (compatible) characters to edges 28

Phylogeny with Distance Matrix 2/27/2021 29

Phylogeny with Distance Matrix n n 2/27/2021 Input is a distance matrix (square, symmetric) between all pair of objects, instead of character state matrix Output is phylogeny with leaves as objects and arcs have distances as labels 30

Phylogeny with Distance Matrix n n n 2/27/2021 Additive matrix: when you can draw a tree where distance between every pair of leaves on the tree is the real distance on distance matrix Matrices are unlikely to be additive in practice For non-additive matrix, minimize deviation over the tree: NP-hard problem 31

Phylogeny with Distance Matrix n n Typically we have 2 matrices: (1) upper bound on distances, and (2) that for lower bounds Metric space: dij>0, dii=0, dij=dji, for all i, j u dij =< dik + dkj u n 2/27/2021 Additive metric spaces follow 4 point condition: dij+dkl=dik+djl >= dil+djk 32

Phylogeny with Distance Matrix n n 2/27/2021 Tree should have 3 -degree internal nodes (Fig 6. 9, p 194) Arc xy to be split proportionately at c, to add a node z by arc cz, so that distances xz, zy are proper 33

Phylogeny with Distance Matrix n n n 2/27/2021 Mxz = dxc + dzc Myz = dyc + dzc Mxy = dxc + dyc Three equations, three unknowns dxc, dyc, dzc to be solved for The tree drawn is unique for 3 objects x, y and z 34

Phylogeny with Distance Matrix n n 2/27/2021 Adding 4 th object w is same as adding 3 rd object z: Add between older objects x and y splitting xy at c 2 If c 2 coincides with c, ignore this and redo the same between zc Object w may hang (from c 2) between xz or yz, but will not have 2 different opportunities 35

Phylogeny with Distance Matrix n n 2/27/2021 The property of uniqueness of the tree remain valid for any k objects for k>4, for metric additive distance matrix The algorithm may have to try all possible places to split an arc, but there will be a unique position, for metric additive space 36

Phylogeny: Ultrametric tree n n 2/27/2021 Excercise: Get MST of a complete graph over table 6. 5 p 195 Ultrametric tree construction: Input: Distance matrices for High cut-off Mh, Low cut-off Ml (table 6. 6 p 201) Output: Phylogeny where leaf-to-leaf distances are within the bounds provided by the 2 matrices (fig 6. 16 p 202) 37

Phylogeny: Ultrametric tree n n 2/27/2021 Algorithm: Compute MST T over Mh (algorithm? ): provides basis for structure of the tree Compute “cut-off” values between each edge on T using Ml: provides basis for distances on the tree edges Compute the ultrametric tree U and find distance on each arc using the cut-offs 38

Phylogeny: Ultrametric tree n n n 2/27/2021 Step 2. 1: input T, output is rooted tree R where internal nodes represent edges of T Sort MST T by edge weights (from Mh) nonincreasing Pick up edges by the sort as root in each iteration The path between the end nodes must go via the root: the two nodes edge should be in two different subtrees Next edge in the sort to be picked up that has the corresponding node (x) on the respective side of the previous root (xy) Until no edge for a node (x) is left (all such xy is picked up), then the node x is on a leaf 39

Phylogeny: Ultrametric tree n n n 2/27/2021 Step 2. 2 (cut-off): For each pair of nodes (x, y) look at the path in R See which is the least common ancestor, say (ab) [note each internal node represents an edge] Look up table Ml, if Ml_xy is more than current cut-off(ab) replace it with M_xy In other words, the highest Ml value on any edge on the path from x to y in T should be its distance on the ultrametric tree On example p 201 -202: root (ad) is updated for pairs of all nodes on the opposite sides EB(1), ED(1), AD(4), AB(3), CB(4), CD(3) 40

Phylogeny: Ultrametric tree n n n 2/27/2021 Step 3 (ultrametric tree): Recompute R again same way as before But, now put distance on internal nodes Height of an internal node is its cut-off / 2 Note, computation of R starts with root downwards Adjust distances between the nodes as heights are being calculated Done 41

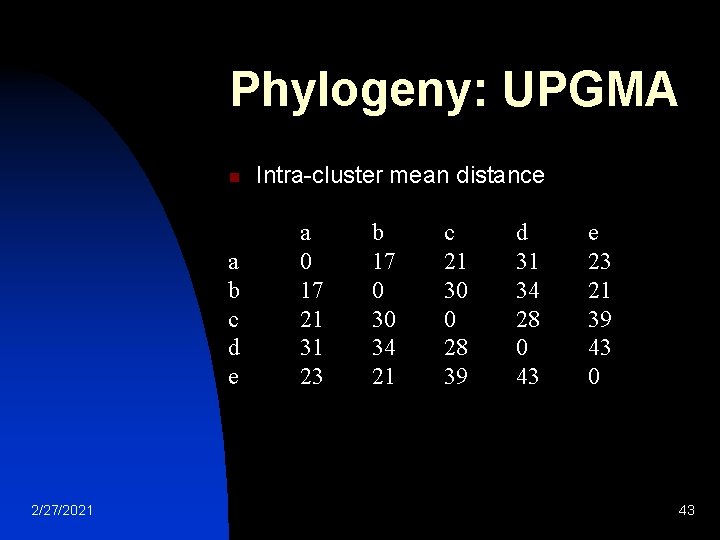
Phylogeny: UPGMA n n n 2/27/2021 Intra-cluster mean distance: Inter-cluster distance WPGMA: distances from the root to every branch tip are equal 42

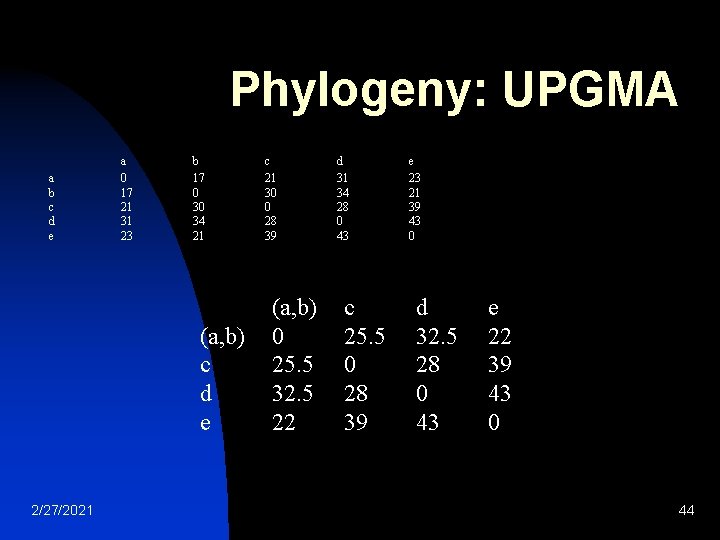
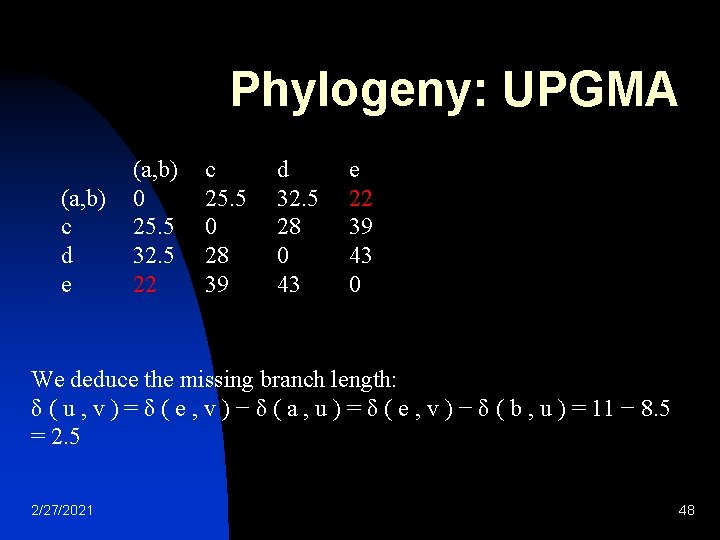
Phylogeny: UPGMA n a b c d e 2/27/2021 Intra-cluster mean distance a 0 17 21 31 23 b 17 0 30 34 21 c 21 30 0 28 39 d 31 34 28 0 43 e 23 21 39 43 0 43

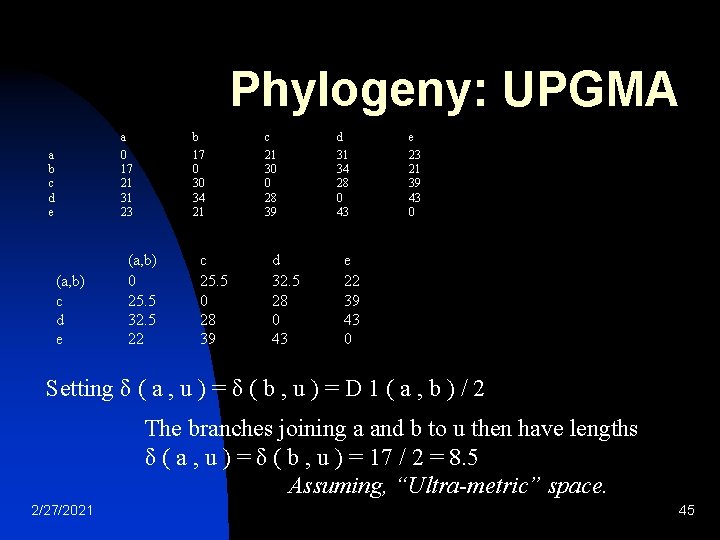
Phylogeny: UPGMA a b c d e a 0 17 21 31 23 b 17 0 30 34 21 (a, b) c d e 2/27/2021 c 21 30 0 28 39 (a, b) 0 25. 5 32. 5 22 d 31 34 28 0 43 c 25. 5 0 28 39 e 23 21 39 43 0 d 32. 5 28 0 43 e 22 39 43 0 44

Phylogeny: UPGMA a 0 17 21 31 23 a b c d e (a, b) c d e b 17 0 30 34 21 (a, b) 0 25. 5 32. 5 22 c 25. 5 0 28 39 c 21 30 0 28 39 d 32. 5 28 0 43 d 31 34 28 0 43 e 23 21 39 43 0 e 22 39 43 0 Setting δ ( a , u ) = δ ( b , u ) = D 1 ( a , b ) / 2 The branches joining a and b to u then have lengths δ ( a , u ) = δ ( b , u ) = 17 / 2 = 8. 5 Assuming, “Ultra-metric” space. 2/27/2021 45

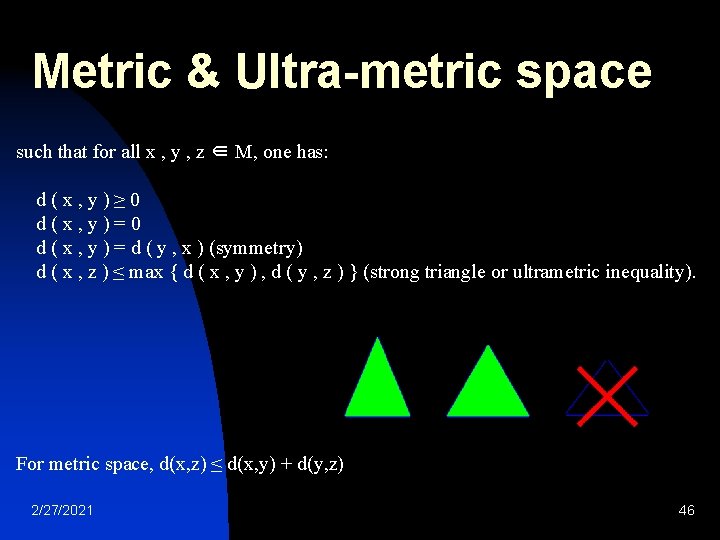
Metric & Ultra-metric space such that for all x , y , z ∈ M, one has: d(x, y)≥ 0 d(x, y)=0 d ( x , y ) = d ( y , x ) (symmetry) d ( x , z ) ≤ max { d ( x , y ) , d ( y , z ) } (strong triangle or ultrametric inequality). For metric space, d(x, z) ≤ d(x, y) + d(y, z) 2/27/2021 46


Phylogeny: UPGMA (a, b) c d e (a, b) 0 25. 5 32. 5 22 c 25. 5 0 28 39 d 32. 5 28 0 43 e 22 39 43 0 We deduce the missing branch length: δ ( u , v ) = δ ( e , v ) − δ ( a , u ) = δ ( e , v ) − δ ( b , u ) = 11 − 8. 5 = 2. 5 2/27/2021 48

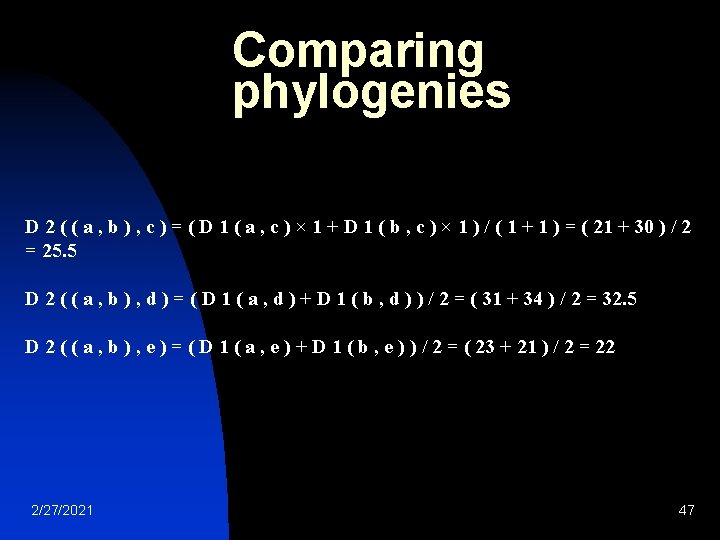
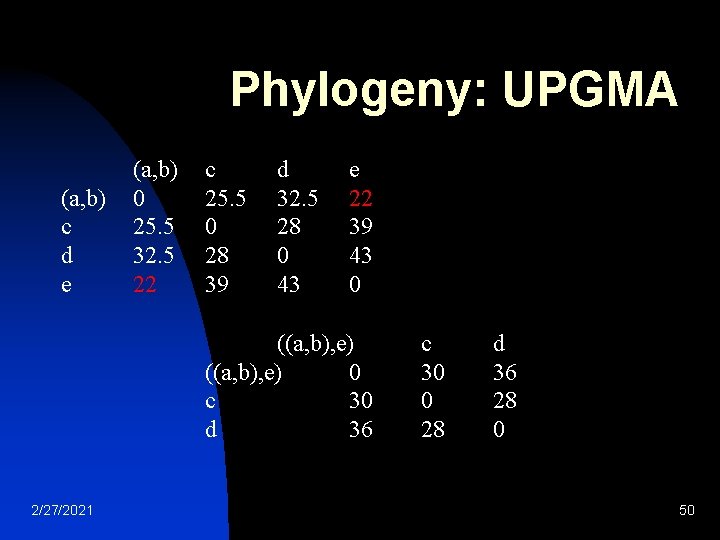
Phylogeny: UPGMA calculated by proportional averaging: D 3 ( ( ( a , b ) , e ) , c ) = ( D 2 ( ( a , b ) , c ) × 2 + D 2 ( e , c ) × 1 ) / ( 2 + 1 ) = ( 25. 5 × 2 + 39 × 1 ) / 3 = 30 Thanks to this proportional average, the calculation of this new distance accounts for the larger size of the ( a , b ) cluster (two elements) with respect to e (one element). Similarly: D 3 ( ( ( a , b ) , e ) , d ) = ( D 2 ( ( a , b ) , d ) × 2 + D 2 ( e , d ) × 1 ) / ( 2 + 1 ) = ( 32. 5 × 2 + 43 × 1 ) / 3 = 36 Replace <proportional average> with mean, you get WPGMA 2/27/2021 49

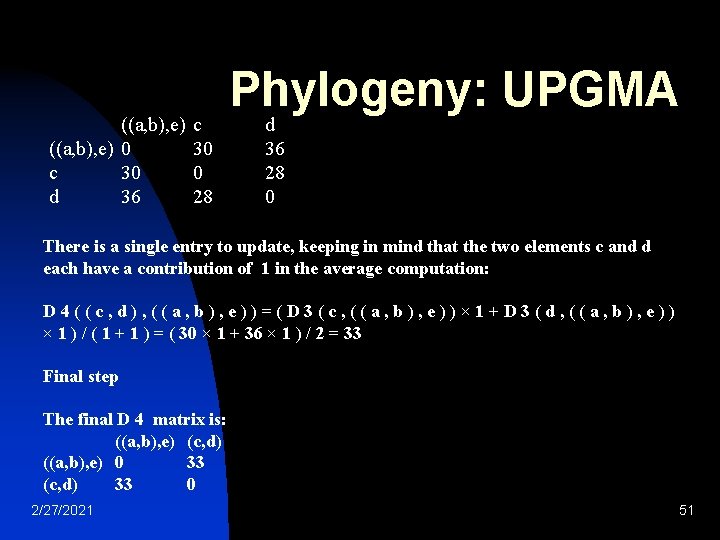
Phylogeny: UPGMA (a, b) c d e (a, b) 0 25. 5 32. 5 22 c 25. 5 0 28 39 d 32. 5 28 0 43 e 22 39 43 0 ((a, b), e) 0 c 30 d 36 2/27/2021 c 30 0 28 d 36 28 0 50

((a, b), e) 0 c 30 d 36 c 30 0 28 Phylogeny: UPGMA d 36 28 0 There is a single entry to update, keeping in mind that the two elements c and d each have a contribution of 1 in the average computation: D 4((c, d), ((a, b), e))=(D 3(c, ((a, b), e))× 1+D 3(d, ((a, b), e)) × 1 ) / ( 1 + 1 ) = ( 30 × 1 + 36 × 1 ) / 2 = 33 Final step The final D 4 matrix is: ((a, b), e) (c, d) ((a, b), e) 0 33 (c, d) 33 0 2/27/2021 51

Phylogeny: UPGMA 2/27/2021 52

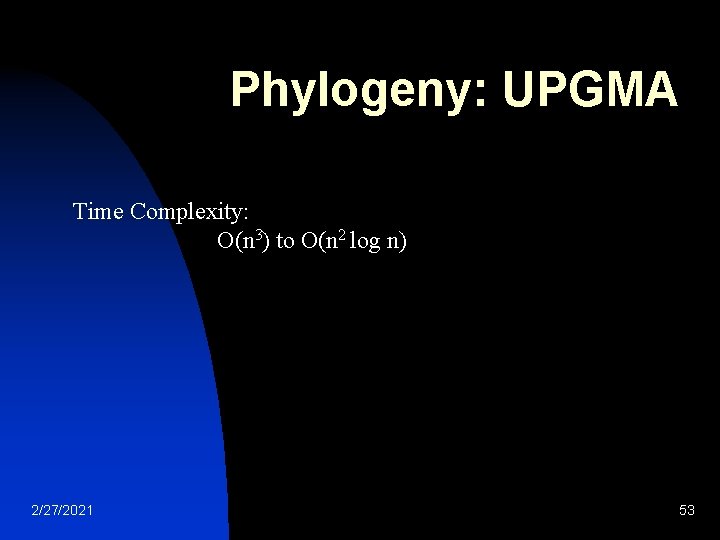
Phylogeny: UPGMA Time Complexity: O(n 3) to O(n 2 log n) 2/27/2021 53

Phylogeny: Neighborjoining • • • Bottom up, as for UPGMA, but non-rooted Distance matrix transformed to Q-matrix (negative) Min Q-value used to connect cluster pairs Cluster distance update formula (in distance-space not Q-space) Iterate: d Q cluster_join d_update iterate • • • Topology: additive distances, node-pair distances are conserved Assumption: “Balanced Minimum Evolution” Greedy optimization (underlying linear programming) “Fast” (? ), O(n 3) complexity for n nodes “Correct” optimized tree even if the source d-matrix is noisy Wiki: https: //en. wikipedia. org/wiki/Neighbor_joining 2/27/2021 54

Phylogeny: General Comments • • • 2/27/2021 Morphological and molecular (sequences) Distance matrix Maximum parsimony Maximum likelihood and Bayesian inference Post-analysis of Tree-support evaluation Shortcomings: convergent evolution, horizontal gene-transfer hybrids, or non-binary tree, or phylogeny network missing species/taxa https: //en. wikipedia. org/wiki/Computational_phylogenetics 55

Comparing phylogenies n n n 2/27/2021 Two trees are expected to be isomorphic All nodes should be on the leaves, if not make it so Pick up a node u and its sibling v on T 1 Look for u in T 2 and if its sibling is not v: return False If the sibling is v then merge uv into its parent (and remove subtree with u and v) Continue bottom up until both T 1 and T 2 become single node trees, then return True 56