6 Protein classifications Domains are reused and reinvented




- Slides: 28

6. Protein classifications



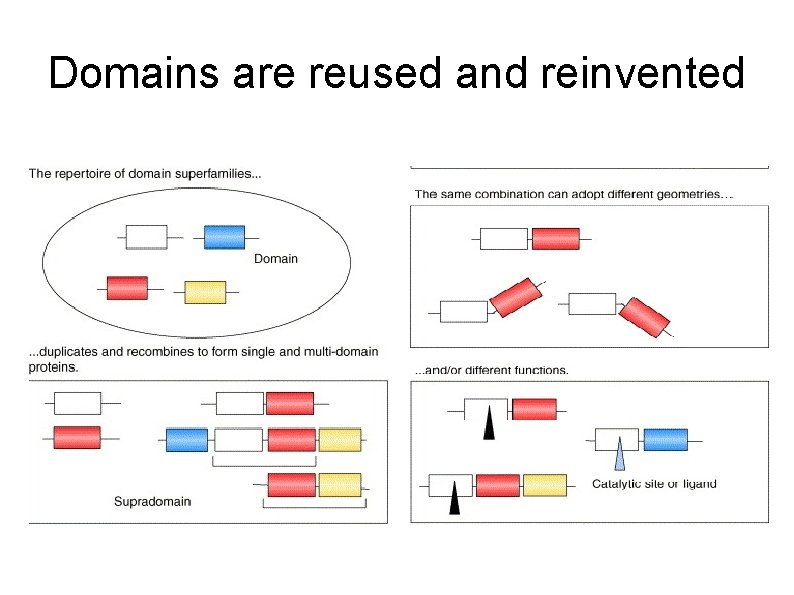
Domains are reused and reinvented

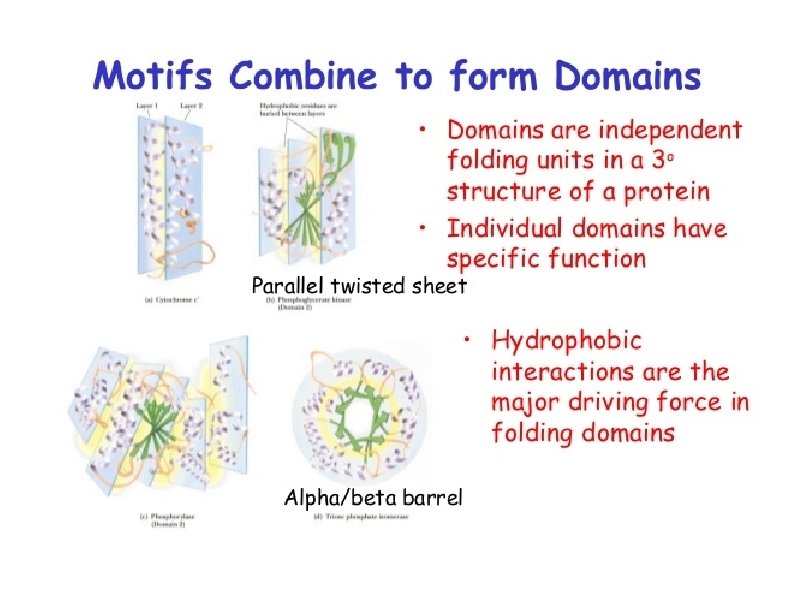
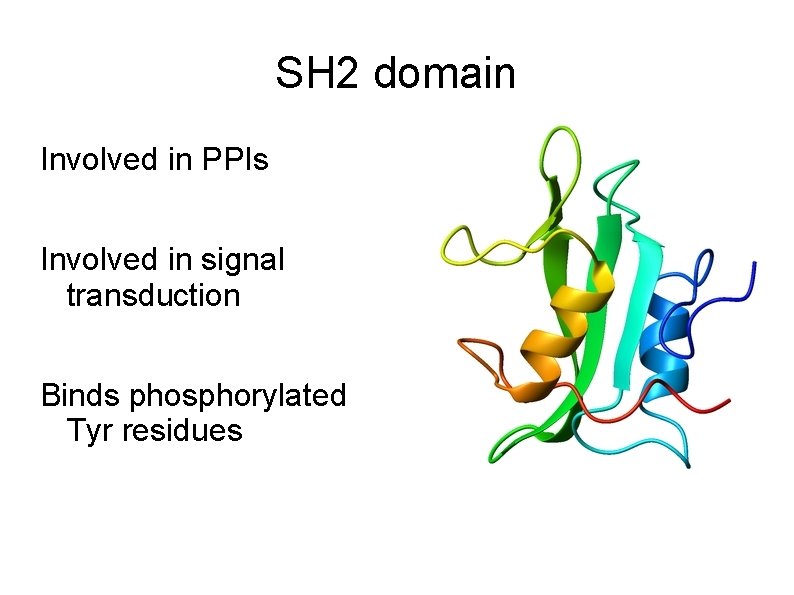
SH 2 domain Involved in PPIs Involved in signal transduction Binds phosphorylated Tyr residues

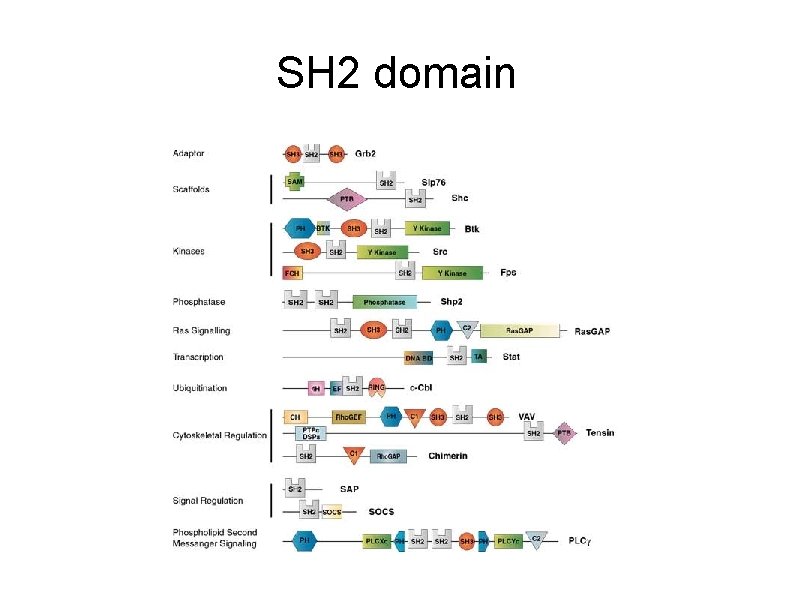
SH 2 domain

Protein classification • Structural domains • Protein families • Sequence signatures

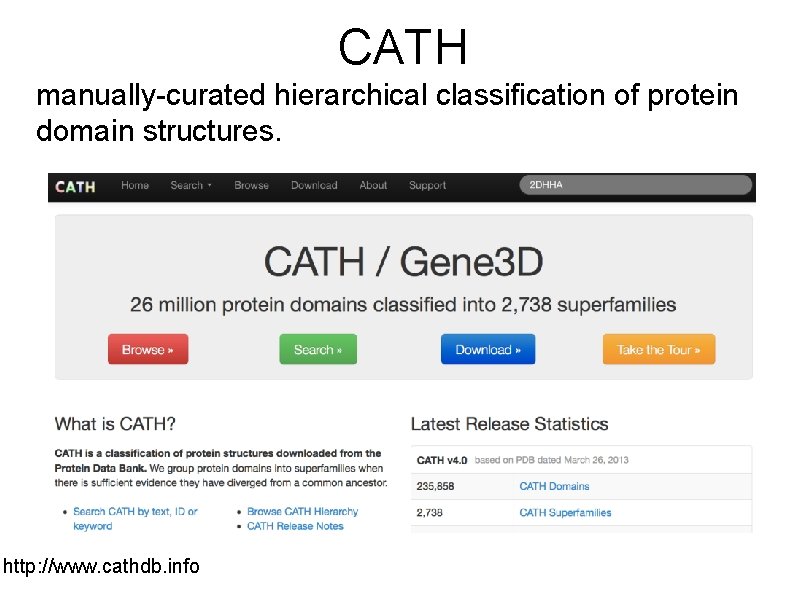
CATH manually-curated hierarchical classification of protein domain structures. http: //www. cathdb. info

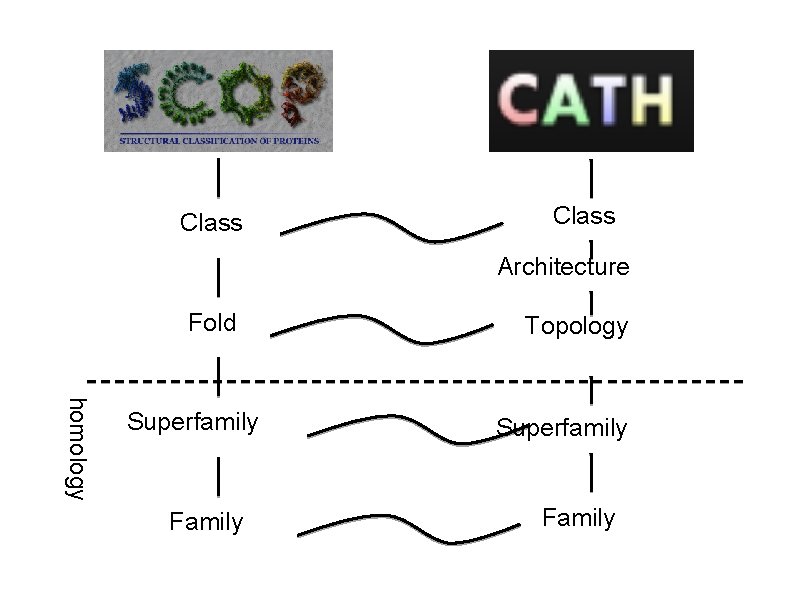
Class: secondary structure content (e. g. mainlyalpha, mainly-beta, mixed alpha/beta or 'few secondary structures'); Architecture: general arrangement of the secondary structures Topology (fold) takes into account the connectivity of secondary structures in the chain; Homologous Superfamily: domains that are believed to be related by a common ancestor.

Class Architecture Fold homology Superfamily Family Topology Superfamily Family

Protein sequence families

Protein families • Defined based on MSA • Identification of functional amino acids • Diversity -> detecting remote homologues • Identification of parts of the sequence space which have no functional annotation

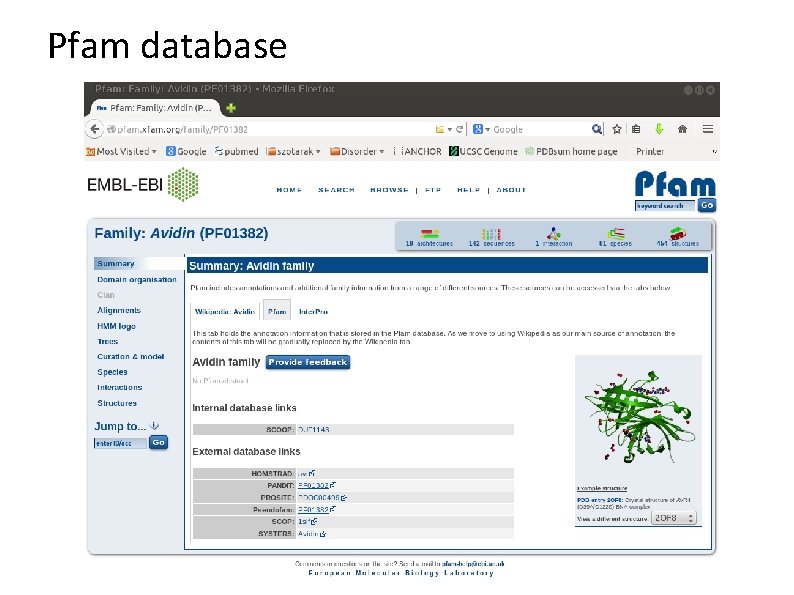
Pfam database

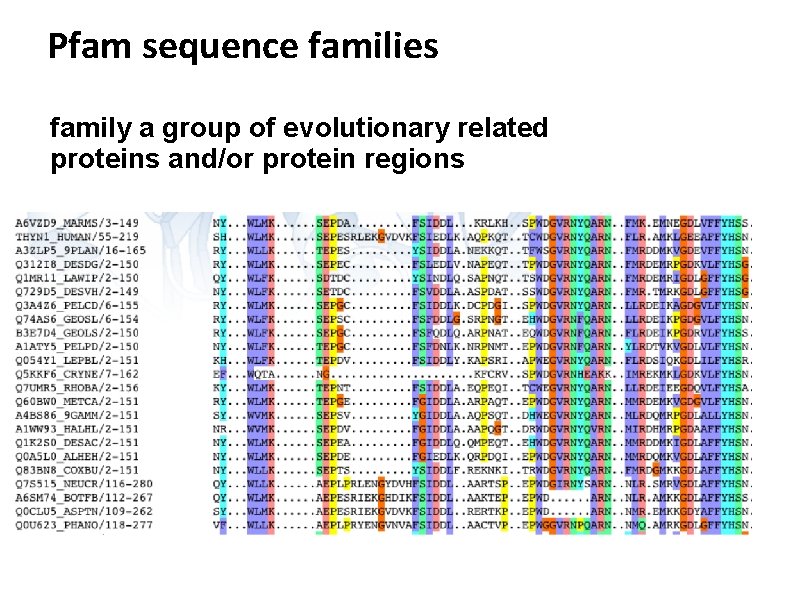
Pfam sequence families family a group of evolutionary related proteins and/or protein regions

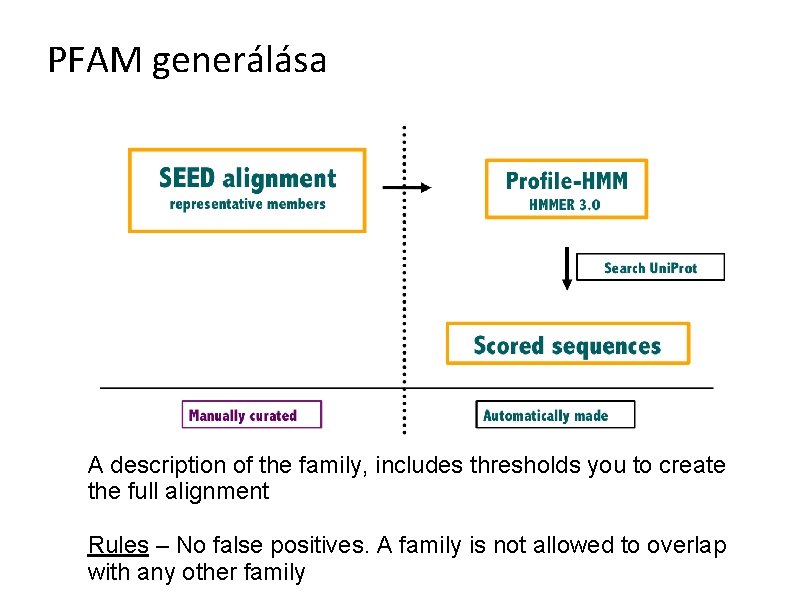
PFAM generálása A description of the family, includes thresholds you to create the full alignment Rules – No false positives. A family is not allowed to overlap with any other family

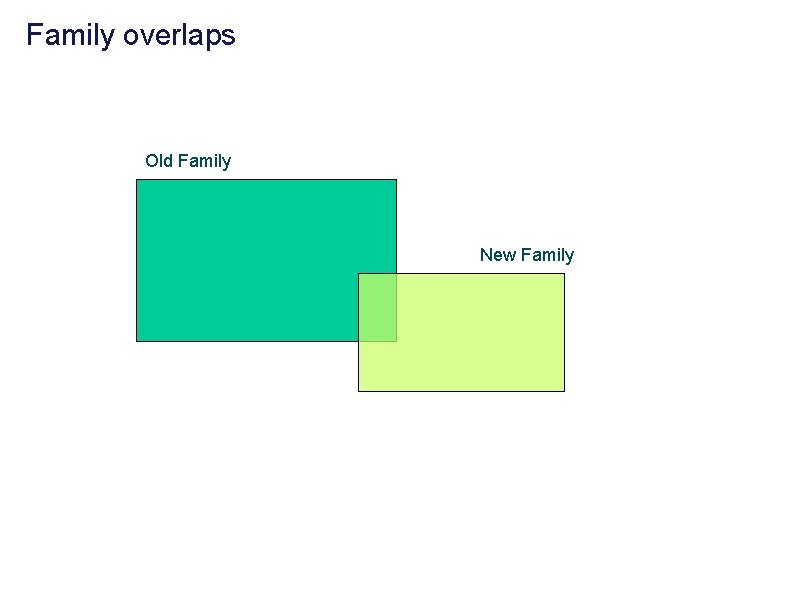
Family overlaps Old Family New Family

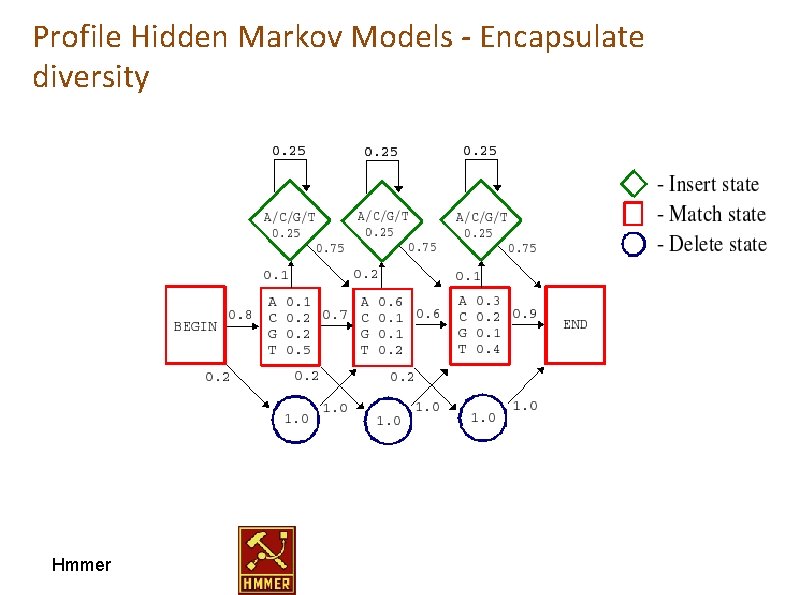
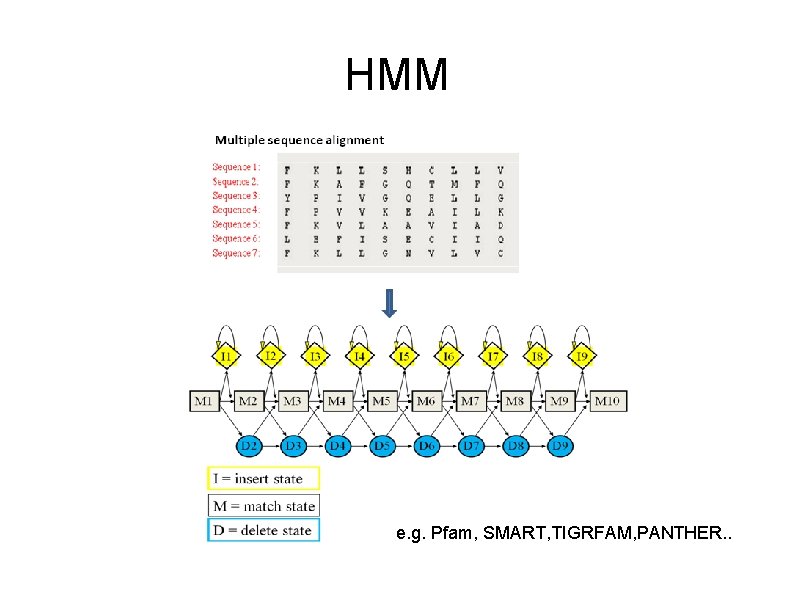
Profile Hidden Markov Models - Encapsulate diversity seq 1 seq 2 seq 3 seq 4 Hmmer ACG-LD SCG--E NCGGFD TCG-WQ

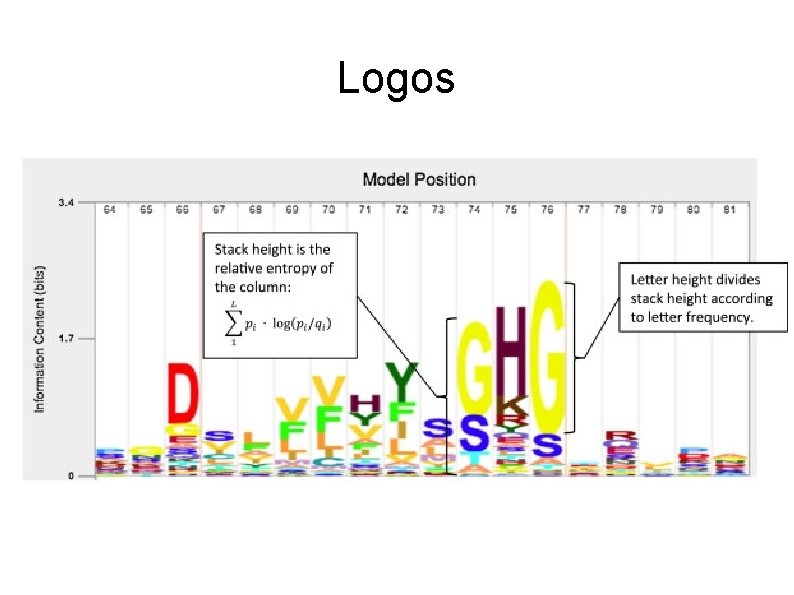
Logos

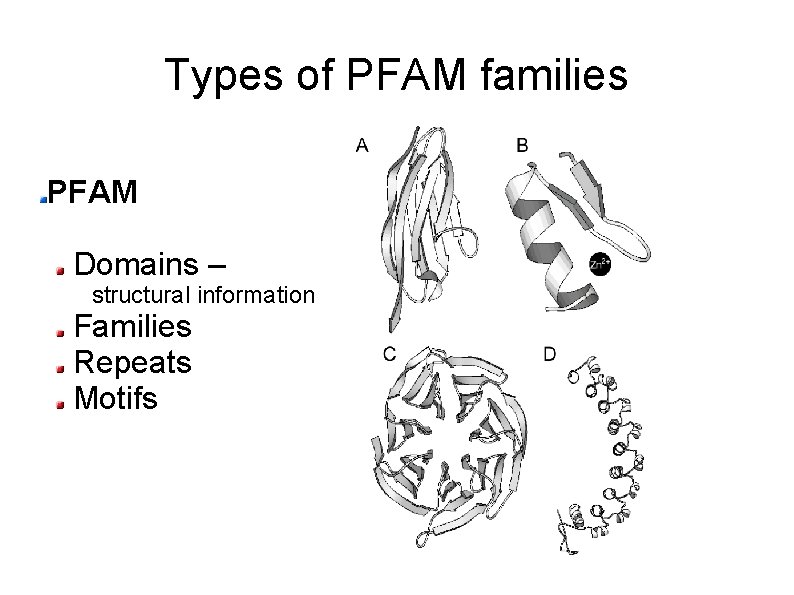
Types of PFAM families PFAM Domains – structural information Families Repeats Motifs

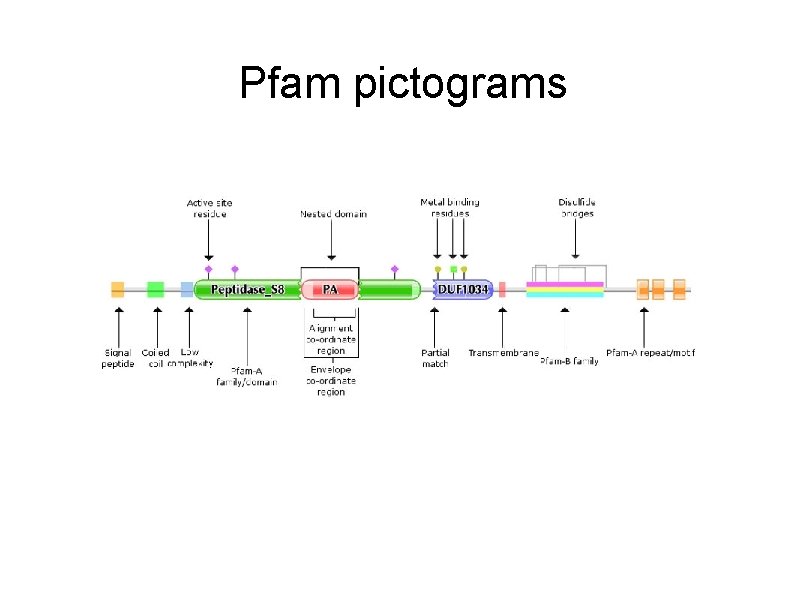
Pfam pictograms

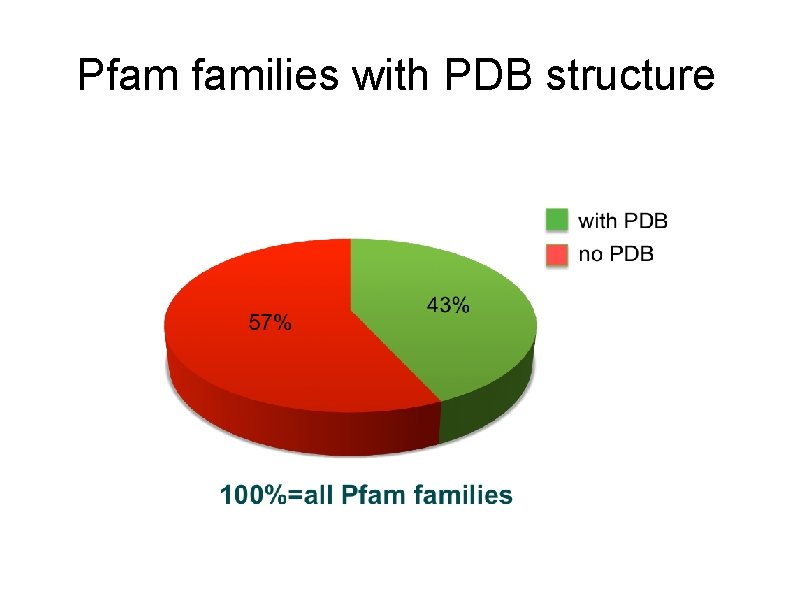
Pfam families with PDB structure

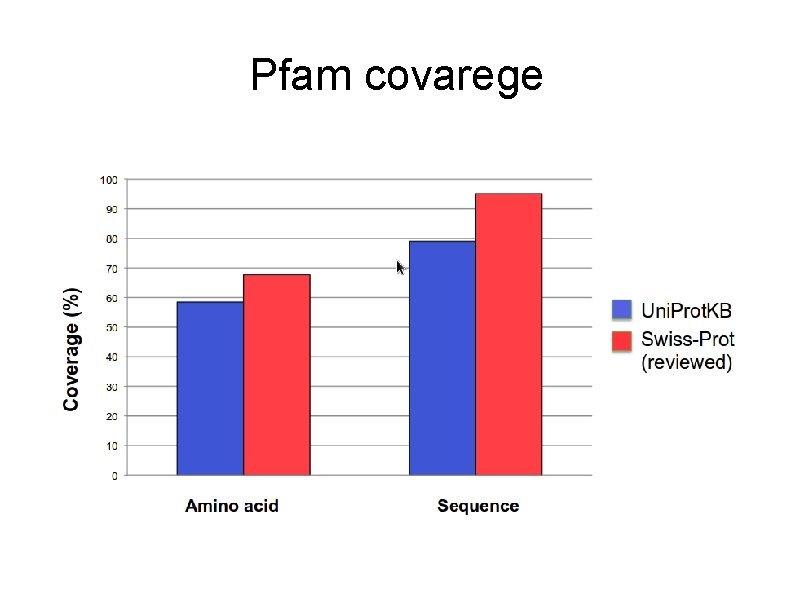
Pfam covarege

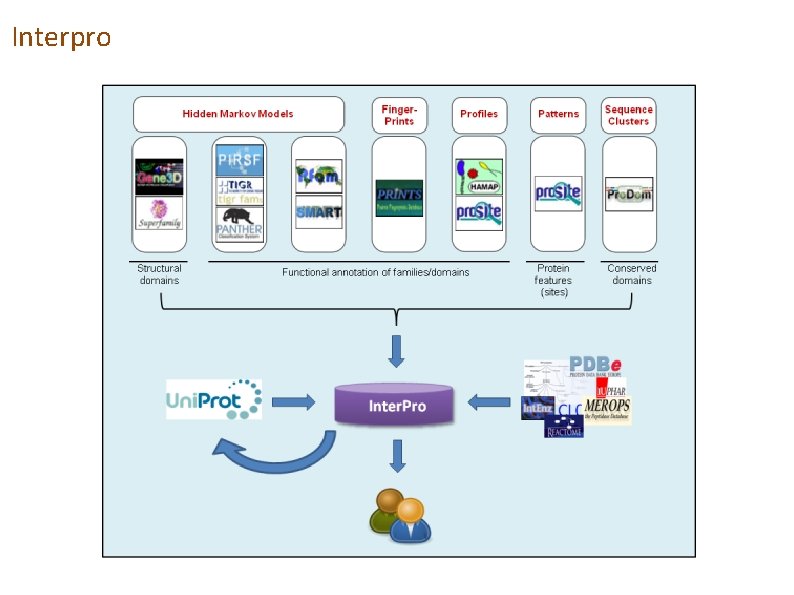
Interpro

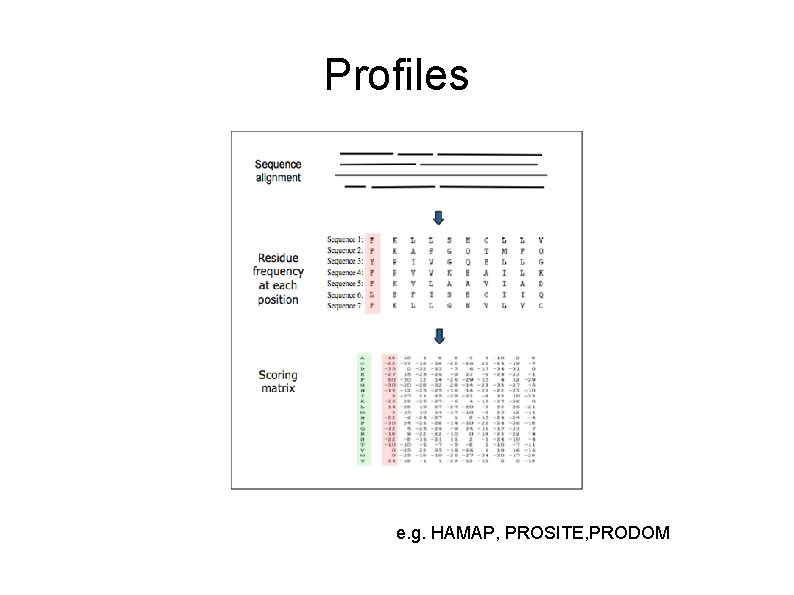
Profiles e. g. HAMAP, PROSITE, PRODOM

HMM e. g. Pfam, SMART, TIGRFAM, PANTHER. .

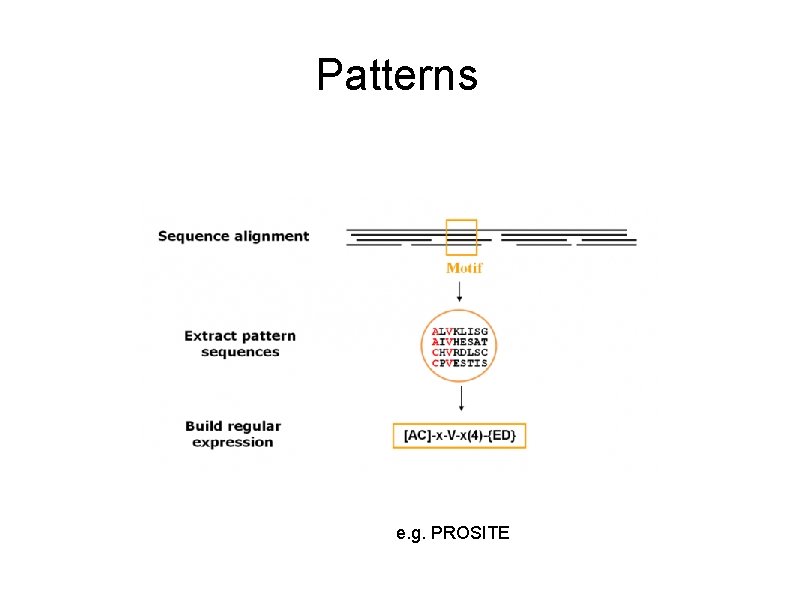
Patterns e. g. PROSITE

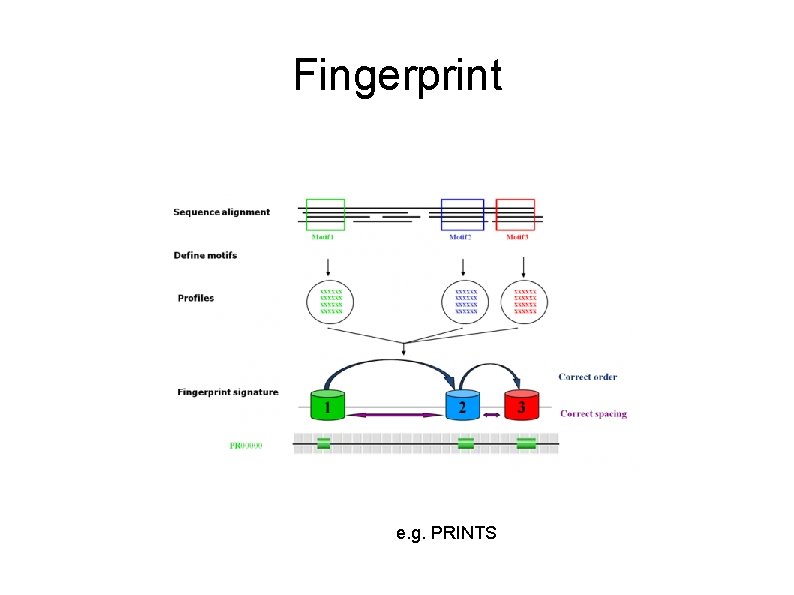
Fingerprint e. g. PRINTS

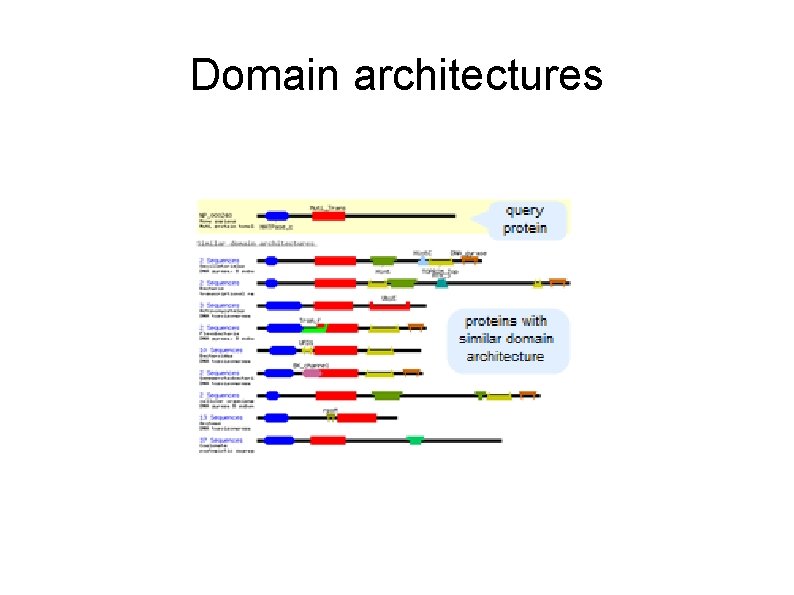
Domain architectures
 Antigentest åre
Antigentest åre Motifs and domains of proteins
Motifs and domains of proteins Jake reinvented quotes
Jake reinvented quotes Netflix reinvented hr
Netflix reinvented hr Jake reinvented chapter summary
Jake reinvented chapter summary Protein pump vs protein channel
Protein pump vs protein channel Protein-protein docking
Protein-protein docking Lightning types and classifications
Lightning types and classifications What are the 11 classification of hand tools
What are the 11 classification of hand tools Cost terms concepts and classifications
Cost terms concepts and classifications Very strongly flavored vegetables
Very strongly flavored vegetables What are the 8 classifications of vegetables?
What are the 8 classifications of vegetables? Mildly flavored vegetables cooking methods
Mildly flavored vegetables cooking methods Rosales classification
Rosales classification Family solanaceae classification
Family solanaceae classification Market forms of mollusks
Market forms of mollusks Classifications of oblique triangles
Classifications of oblique triangles Ct dph reportable event classifications
Ct dph reportable event classifications Molisch test
Molisch test Dewey decimal system example
Dewey decimal system example Draft horse organism classifications
Draft horse organism classifications Unsd classifications
Unsd classifications Anatomy of a fire extinguisher
Anatomy of a fire extinguisher Data center ratings
Data center ratings Asthma classification
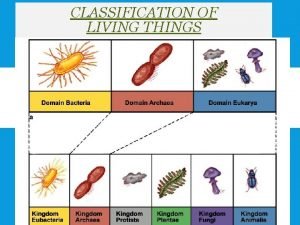
Asthma classification Kingdom archaea
Kingdom archaea Classification of vegetables
Classification of vegetables Key to the world
Key to the world What are the 7 classifications of living things
What are the 7 classifications of living things