6 Kernel Regression Framework Phenotype Genetic Value Model
![RKHS Regressions (Background) - Uses: - Scatter-plot smoothing (Smoothing Splines) [1] - Spatial smoothing RKHS Regressions (Background) - Uses: - Scatter-plot smoothing (Smoothing Splines) [1] - Spatial smoothing](https://slidetodoc.com/presentation_image_h2/8cbedb2feb408f7c8266f20e470c2c5f/image-3.jpg)
![Functions as Gaussian processes K=A => Animal Model [1] de los Campos Gianola and Functions as Gaussian processes K=A => Animal Model [1] de los Campos Gianola and](https://slidetodoc.com/presentation_image_h2/8cbedb2feb408f7c8266f20e470c2c5f/image-5.jpg)

![How to Choose the Reproducing Kernel? [1] Þ Pedigree-models K=A Þ Genomic Models: - How to Choose the Reproducing Kernel? [1] Þ Pedigree-models K=A Þ Genomic Models: -](https://slidetodoc.com/presentation_image_h2/8cbedb2feb408f7c8266f20e470c2c5f/image-9.jpg)


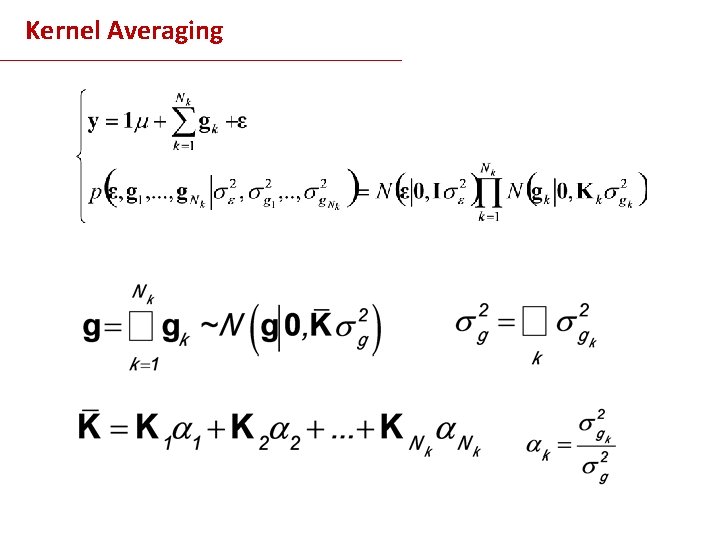



- Slides: 22

6. Kernel Regression

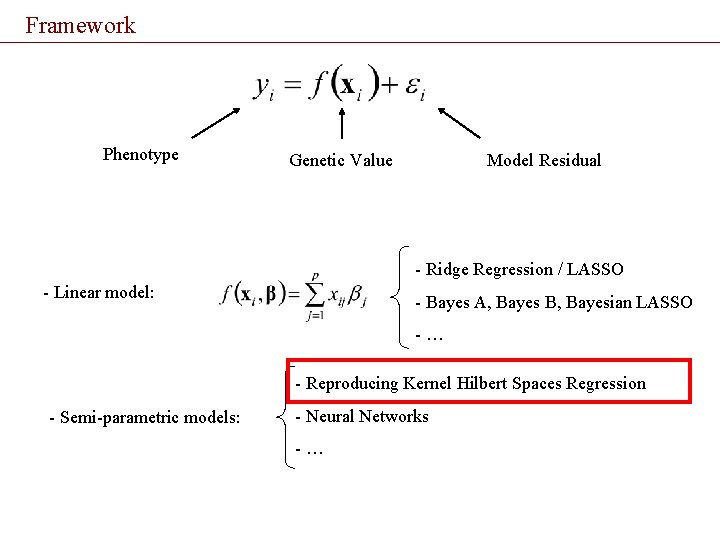
Framework Phenotype Genetic Value Model Residual - Ridge Regression / LASSO - Linear model: - Bayes A, Bayes B, Bayesian LASSO -… - Reproducing Kernel Hilbert Spaces Regression - Semi-parametric models: - Neural Networks -…
![RKHS Regressions Background Uses Scatterplot smoothing Smoothing Splines 1 Spatial smoothing RKHS Regressions (Background) - Uses: - Scatter-plot smoothing (Smoothing Splines) [1] - Spatial smoothing](https://slidetodoc.com/presentation_image_h2/8cbedb2feb408f7c8266f20e470c2c5f/image-3.jpg)
RKHS Regressions (Background) - Uses: - Scatter-plot smoothing (Smoothing Splines) [1] - Spatial smoothing (‘Kriging’) [2] - Classification problems (Support vector machines) [3] - Animal model -… - Regression setting [1] Wahba (1990) Spline Models for Observational Data. [2] Cressie, N. (1993) Statistics for Spatial Data. [3] Vapnik, V. (1998) Statistical Learning Theory. - (it can be of any nature) - unknown function

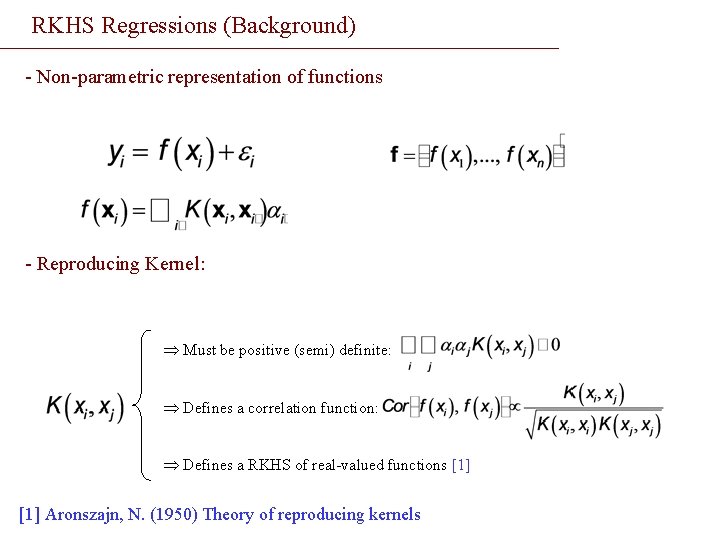
RKHS Regressions (Background) - Non-parametric representation of functions - Reproducing Kernel: Þ Must be positive (semi) definite: Þ Defines a correlation function: Þ Defines a RKHS of real-valued functions [1] Aronszajn, N. (1950) Theory of reproducing kernels
![Functions as Gaussian processes KA Animal Model 1 de los Campos Gianola and Functions as Gaussian processes K=A => Animal Model [1] de los Campos Gianola and](https://slidetodoc.com/presentation_image_h2/8cbedb2feb408f7c8266f20e470c2c5f/image-5.jpg)
Functions as Gaussian processes K=A => Animal Model [1] de los Campos Gianola and Rosa (2008) Journal of Animal Sci.

RKHS Regression in BGLR 1 ETA<-list(K=K, model='RKHS') ) fm<-BGLR(y=y, ETA=ETA, n. Iter=. . . ) [1]: the algorithm is described in de los Campos et al. Genetics Research (2010)

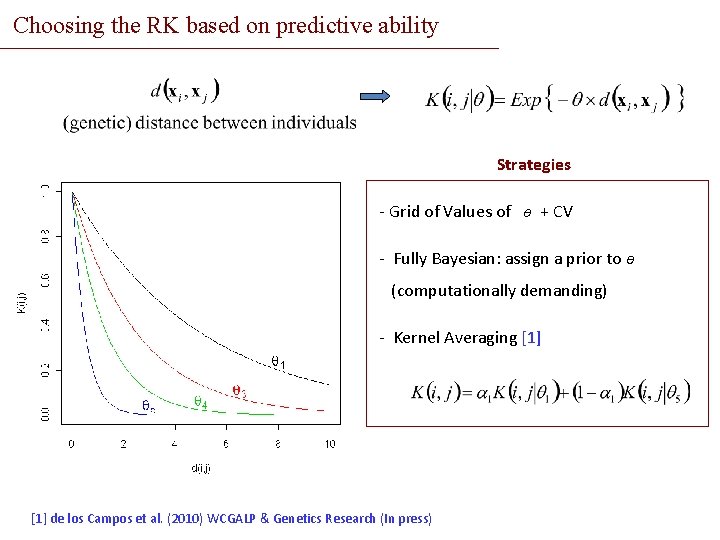
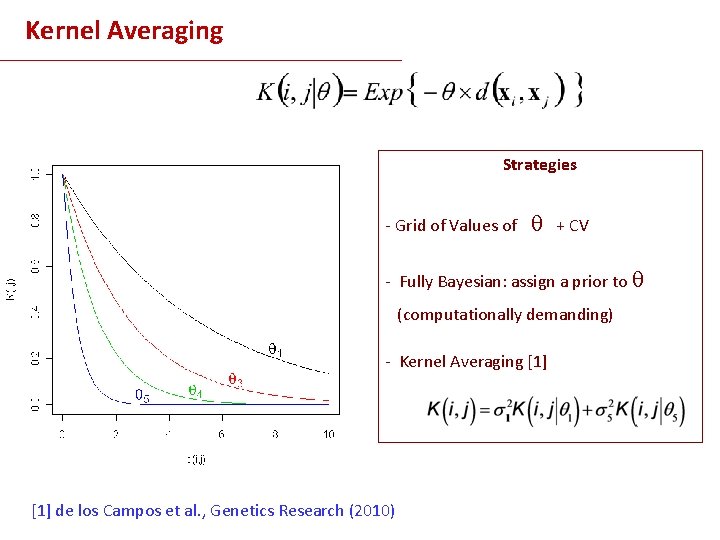
Choosing the RK based on predictive ability Strategies - Grid of Values of ө + CV - Fully Bayesian: assign a prior to ө (computationally demanding) - Kernel Averaging [1] de los Campos et al. (2010) WCGALP & Genetics Research (In press)

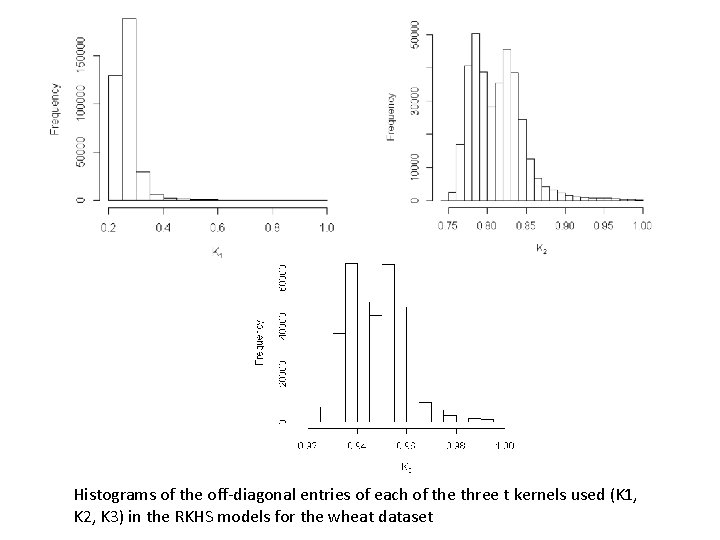
Histograms of the off-diagonal entries of each of the three t kernels used (K 1, K 2, K 3) in the RKHS models for the wheat dataset
![How to Choose the Reproducing Kernel 1 Þ Pedigreemodels KA Þ Genomic Models How to Choose the Reproducing Kernel? [1] Þ Pedigree-models K=A Þ Genomic Models: -](https://slidetodoc.com/presentation_image_h2/8cbedb2feb408f7c8266f20e470c2c5f/image-9.jpg)
How to Choose the Reproducing Kernel? [1] Þ Pedigree-models K=A Þ Genomic Models: - Marker-based kinship Model-derived Kernel Predictive Approach - Explore a wide variety of kernels => Cross-validation => Bayesian methods [1] Shawne-Taylor and Cristianini (2004)

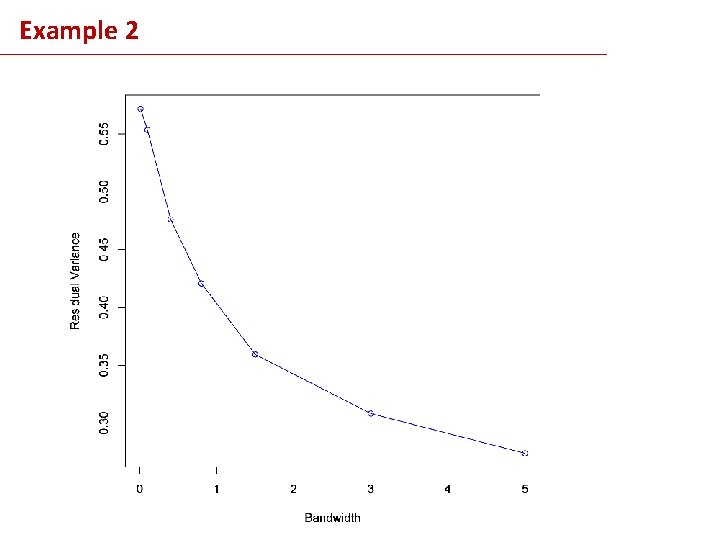
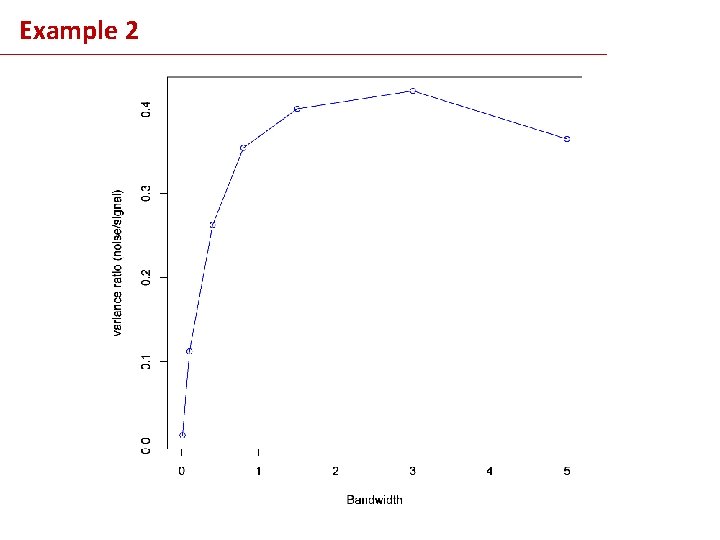
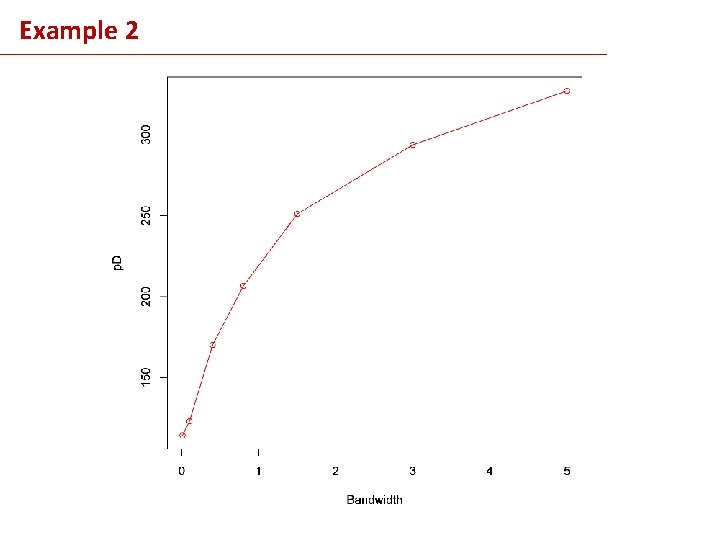
Example 2

Example 2

Example 2

Example 2

Example 3 Kernel Averaging

Kernel Averaging Strategies - Grid of Values of + CV - Fully Bayesian: assign a prior to (computationally demanding) - Kernel Averaging [1] de los Campos et al. , Genetics Research (2010)
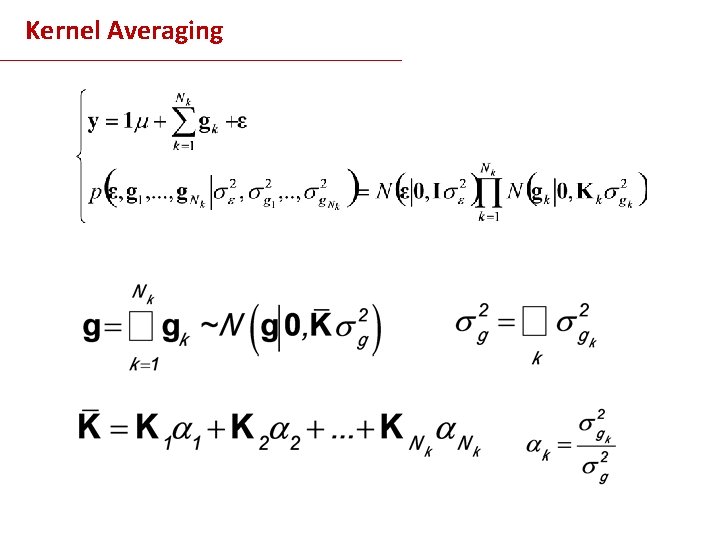
Kernel Averaging

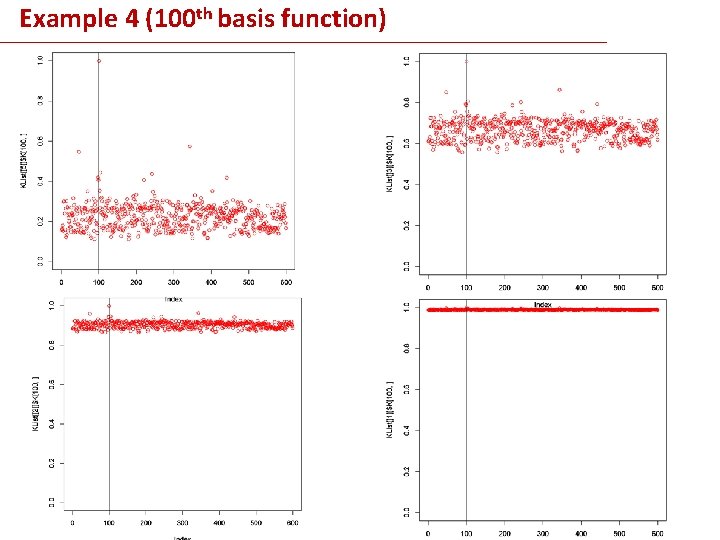
Example 4 (100 th basis function)

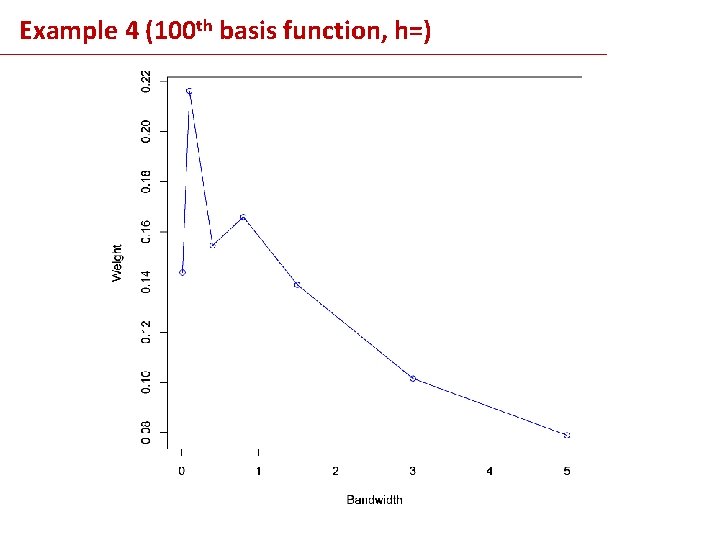
Example 4 (100 th basis function, h=)

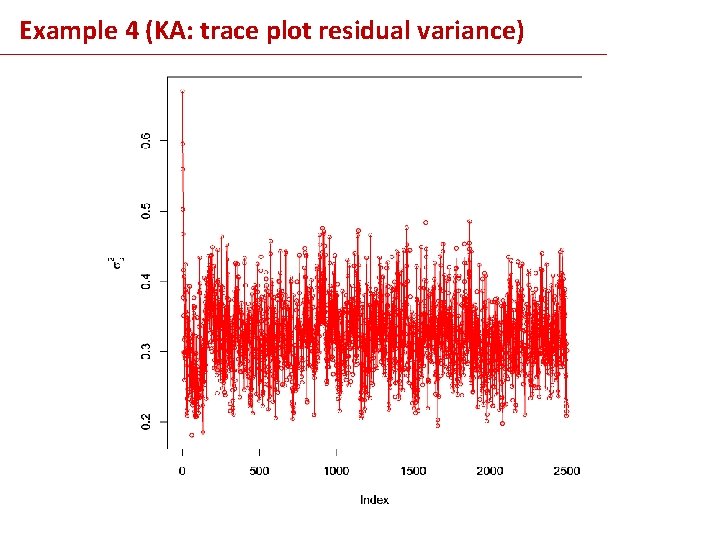
Example 4 (KA: trace plot residual variance)

Example 4 (KA: trace plot kernel-variances)

Example 4 (KA: trace plot kernel-variances)

Example 4 (KA: prediction accuracy)