2 Person Mixture 4 Found Underwear Majorminor Mixture










































- Slides: 82

2 Person Mixture #4 Found Underwear Major/minor Mixture

Scenario • Victim and Accused were both at a party held at a local park • Victim says Accused raped her behind a bush • Underwear found behind bush submitted • Sperm found • Differential extraction wasn’t great

Scenario • Subject says he had nothing to do with anything • But… • He does say that “some girl” masturbated him • So IF his DNA shows up on in a pair of panties or someone’s lady parts… • Maybe “that girl” didn’t wash her hands before she used the restroom • (Actual defense theory proffered to me!)

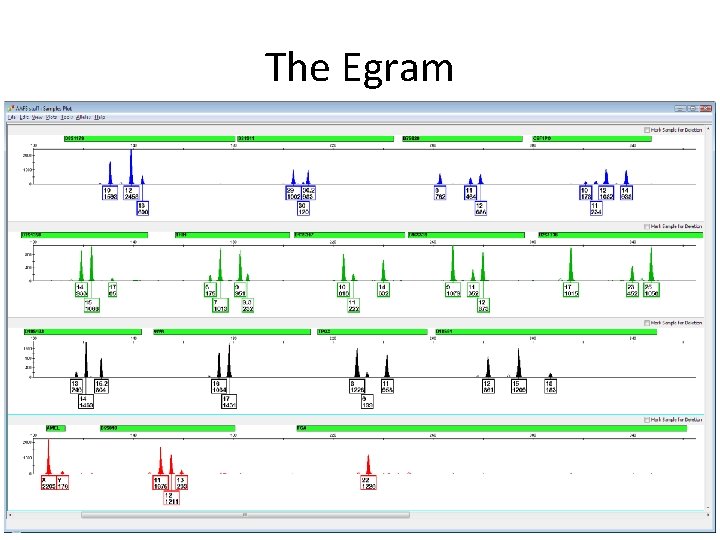
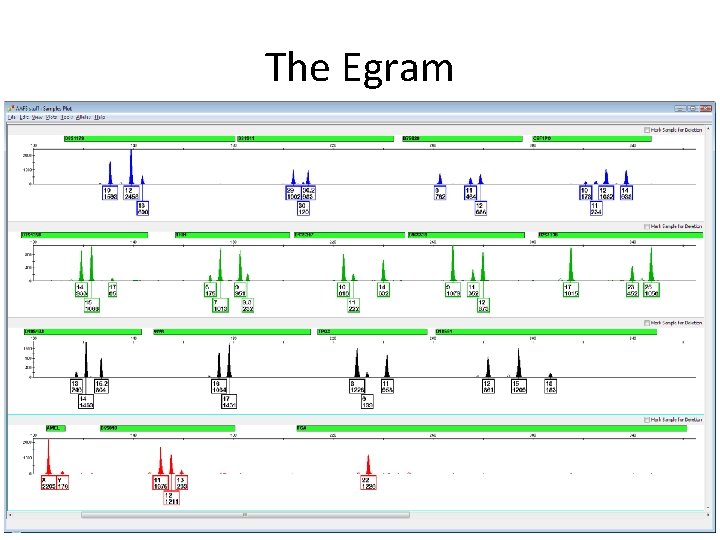
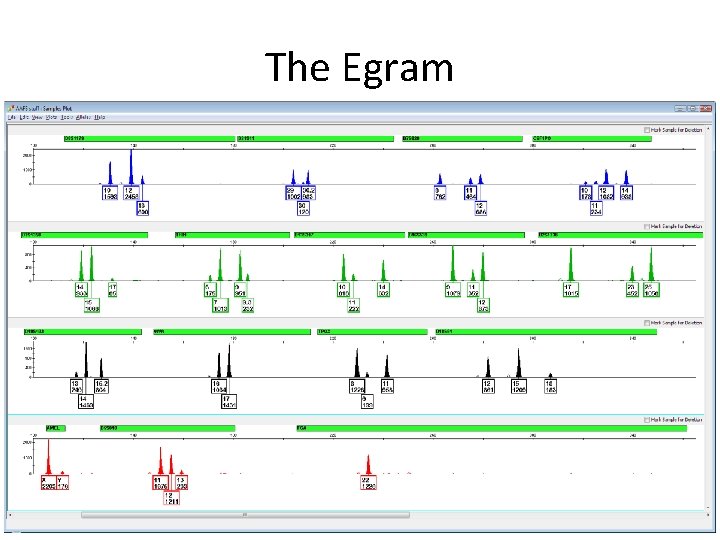
The Egram

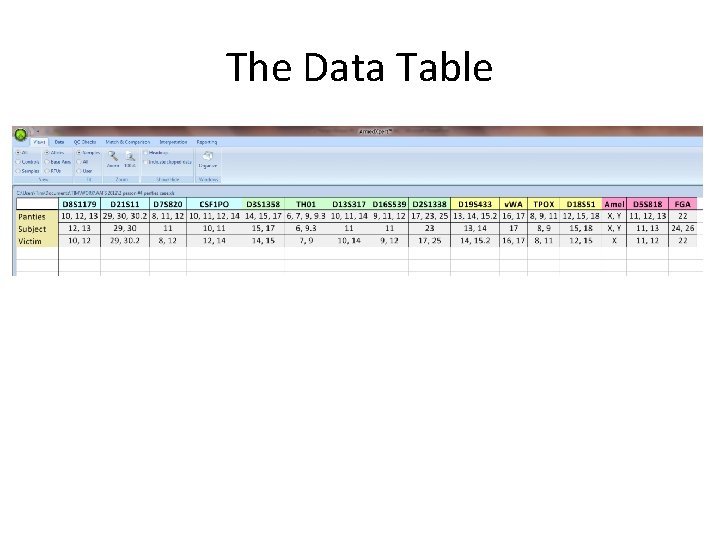
The Data Table

V and S 1. Both included, but that’s all I can do 2. The minor cannot be interpreted (inconclusive) 3. Not sure how many contributors 4. I can separate major from minor (2 stats) 0 of 30 30 Countdown

Interpretation • • • Consistent with 2 people Major/minor (or close enough – D 7) Some minor alleles >300 – restricted RMP Some minor alleles <300 – modified RMP FGA shows only a single allele Cannot assume anyone or anything as far as being a contributor

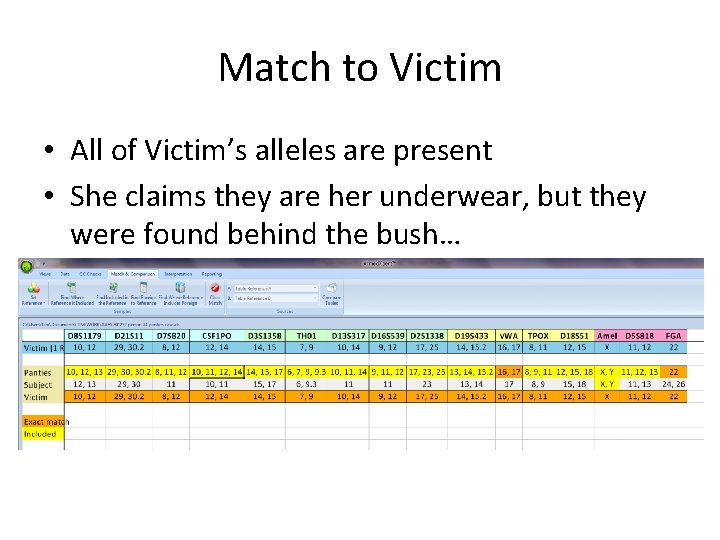
Match to Victim • All of Victim’s alleles are present • She claims they are her underwear, but they were found behind the bush…

Match to Suspect • • Let’s skip this step for now (Don’t consider the references…) This profile may be missing things It looks like a more complicated stat may be coming

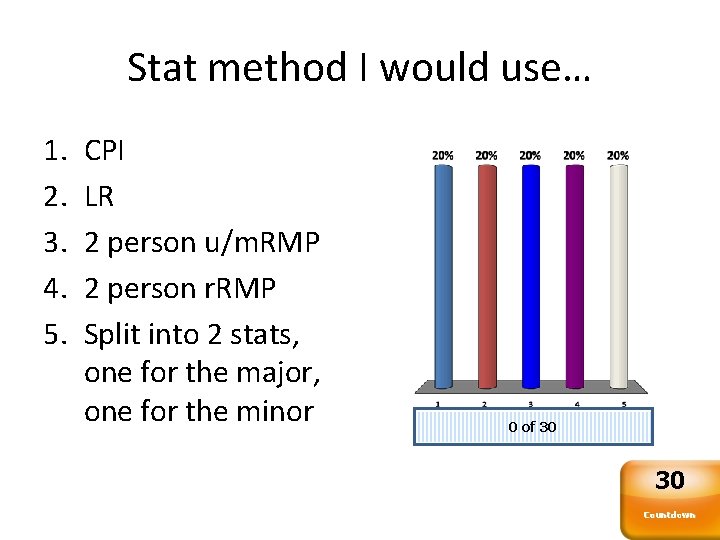
Stat method I would use… 1. 2. 3. 4. 5. CPI LR 2 person u/m. RMP 2 person r. RMP Split into 2 stats, one for the major, one for the minor 0 of 30 30 Countdown

Stat method • We can discuss all 5 of those stat options • Some will have more success than others • Some are (much) more complicated than others

The CPI stat • If it is a mixture of 2 people, why would you use it? (It’s not indeterminate) • You cannot use loci with alleles in the Danger Zone (<300 for this data) • That leaves only 6 loci to use for CPI

The CPI stat • D 8, D 7, D 2 – are OK • D 16 – minor is >300 by a bit but is in stutter position – better check it • v. WA – only 2 alleles, what if minor dropped out? • FGA – only 1 allele, CPI would only account for a single homozygote

The RMP stat • Three options • First the Fast Way – modified RMP • Then the Almost As Fast Way – restricted where we can – modified where we can’t restrict • Then the Slow Way – Major/minor – A stat for each

The Fast Way • Just open the window and click the “Mixture Frequency”

The Fast Way • Then open the Frequency Report • This is a modified RMP – Either Allele, Any – Or restricted RMP

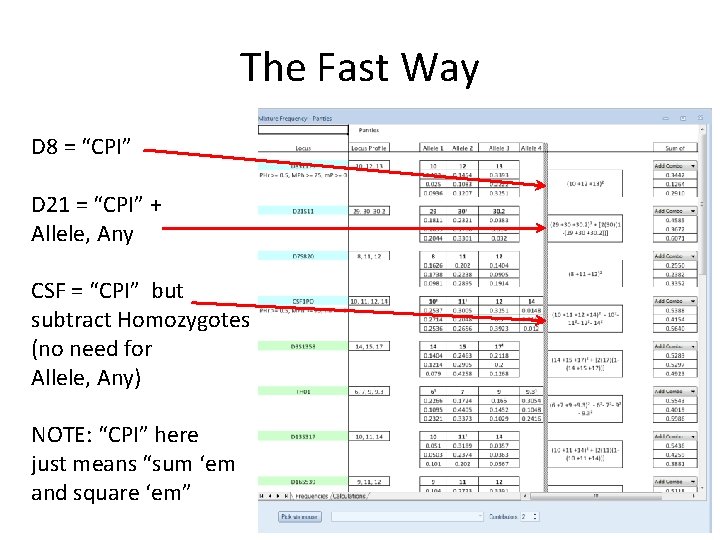
The Fast Way D 8 = “CPI” D 21 = “CPI” + Allele, Any CSF = “CPI” but subtract Homozygotes (no need for Allele, Any) NOTE: “CPI” here just means “sum ‘em and square ‘em”

The Fast Way • If you want to really study a locus, just hover it • This is CSF • Or hit the “Calculations” tab

The Fast Way • Every calculation for every locus is this way • If you copy and paste into Excel you can check the math – Add “=“, change 2 to ^2, and replace []

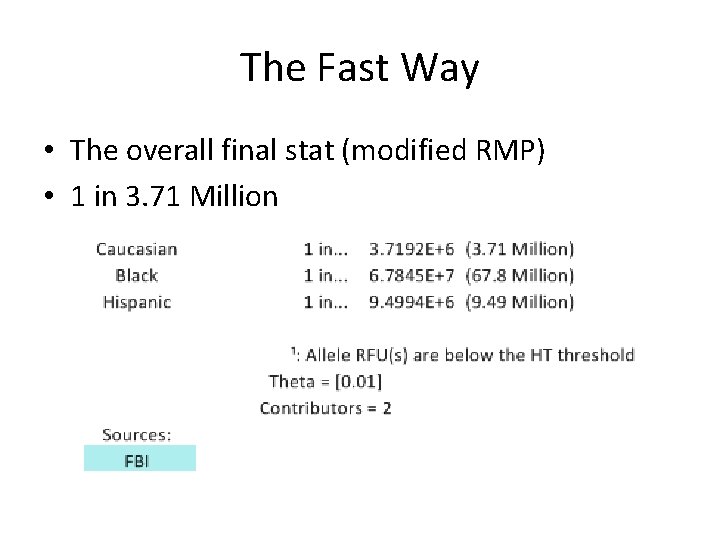
The Fast Way • The overall final stat (modified RMP) • 1 in 3. 71 Million

The Almost as Fast Way • The loci where we’re not concerned about drop out we’ll use restricted rather than modified • Also for any 4 allele loci • Remember, you set the rules here, not the program

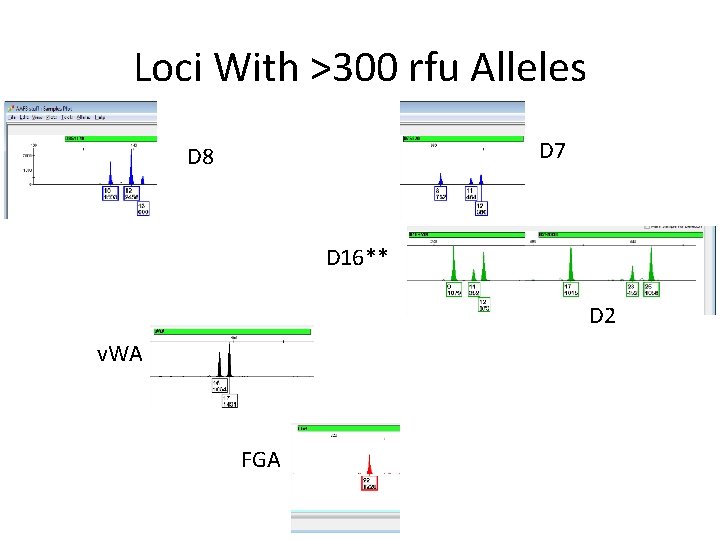
The Egram

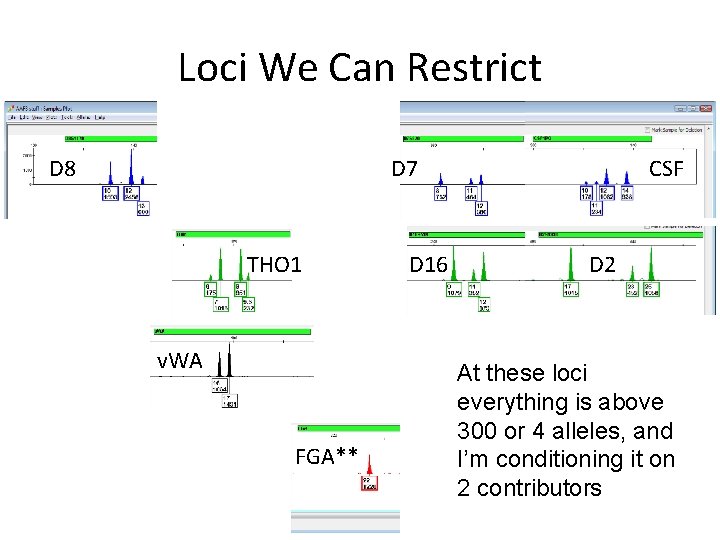
Loci We Can Restrict D 8 D 7 THO 1 v. WA FGA** D 16 CSF D 2 At these loci everything is above 300 or 4 alleles, and I’m conditioning it on 2 contributors

The Almost As Fast Way • So we’ll switch the following loci to restricted RMP: This is very easy: I just tell it to use – – – – CSF restricted for the loci I want in from that “Mixture Frequency” window in THO 1 the Interpretation window D 8 D 7 D 16 D 2 v. WA FGA (well, maybe 22, Any)

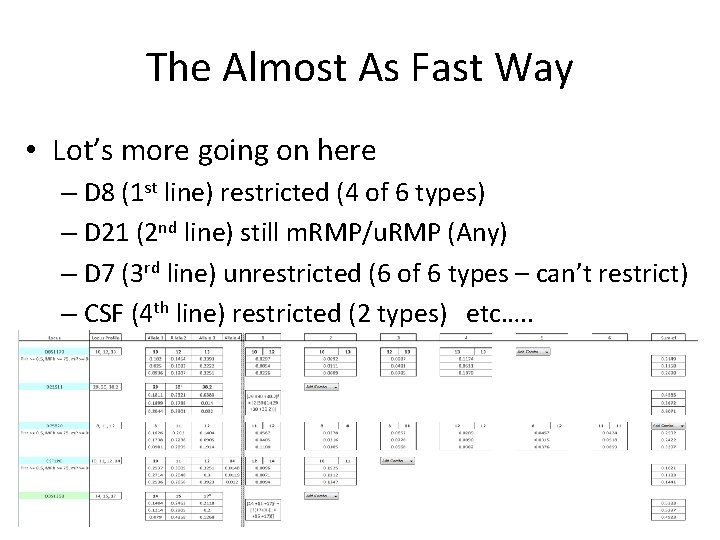
The Almost As Fast Way • Lot’s more going on here – D 8 (1 st line) restricted (4 of 6 types) – D 21 (2 nd line) still m. RMP/u. RMP (Any) – D 7 (3 rd line) unrestricted (6 of 6 types – can’t restrict) – CSF (4 th line) restricted (2 types) etc…. .

The Almost As Fast Way • The overall final stat – About ½ modified RMP – About ½ restricted RMP – Plus Allele, Any at FGA • 1 in 6. 8 million - about twice what it was

The Slow Way • We’ll do what we can to come up with a major and a minor profile • Use the “Popout Calls” feature after hitting “View call report” • This will give us two new profiles we can name Major and Minor • Remember the Egram

The Slow Way • Start with 4 Allele loci • We need to keep track of loci with alleles <300 rfu • Be ready for the “obligate” function and Anys for the minor • I didn’t really figure out a way to do power point slides to show these steps very well • Please bear with me

The Egram

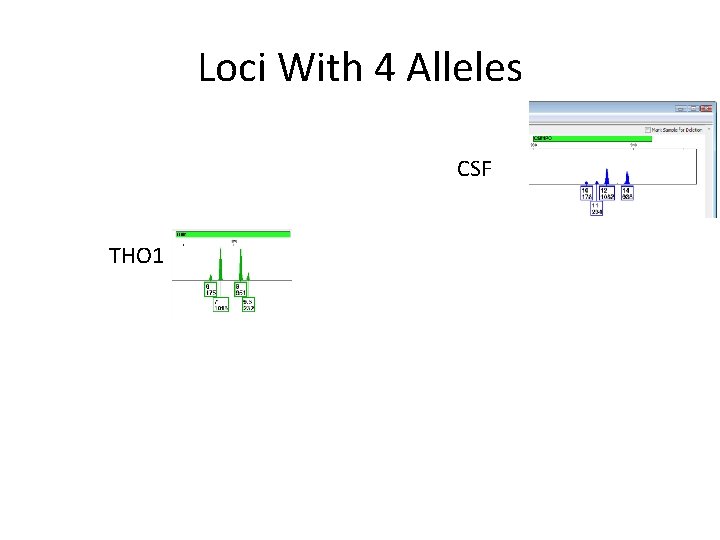
Loci With 4 Alleles CSF THO 1

The Slow Way • • We’ll start with 4 allele loci Most information We can get a “for sure” mixture proportion CSF and THO 1 give us a Major at 83%

The Egram

Loci With >300 rfu Alleles D 7 D 8 D 16** D 2 v. WA FGA

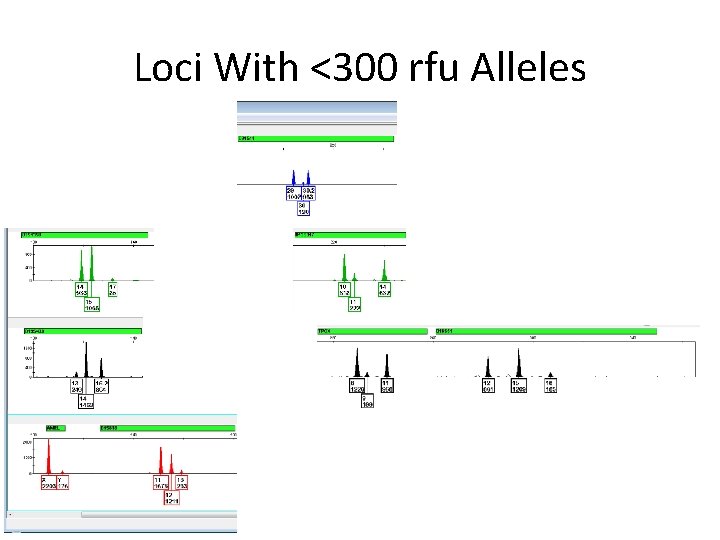
Loci With <300 rfu Alleles

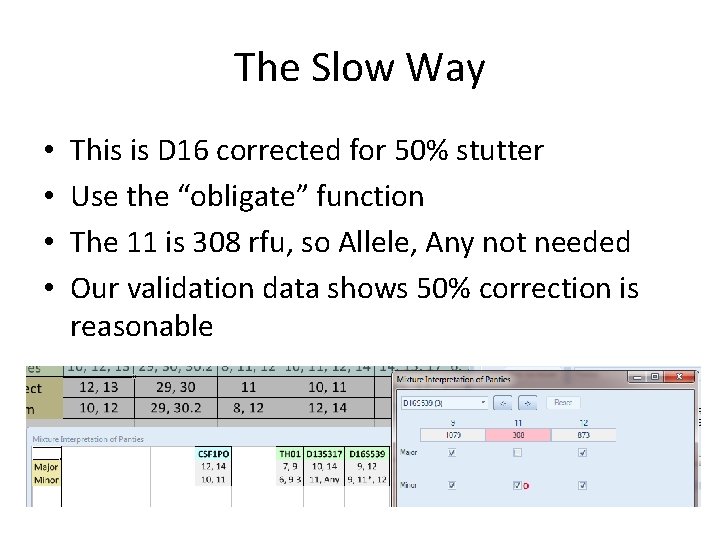
The Slow Way • Remind me to look at D 16 after I finish splitting into major/minor – Minor allele in stutter position – Is it really >300? – Should we use Alelle, Any to be safe? – We need to find out

The Slow Way • • This is D 16 corrected for 50% stutter Use the “obligate” function The 11 is 308 rfu, so Allele, Any not needed Our validation data shows 50% correction is reasonable

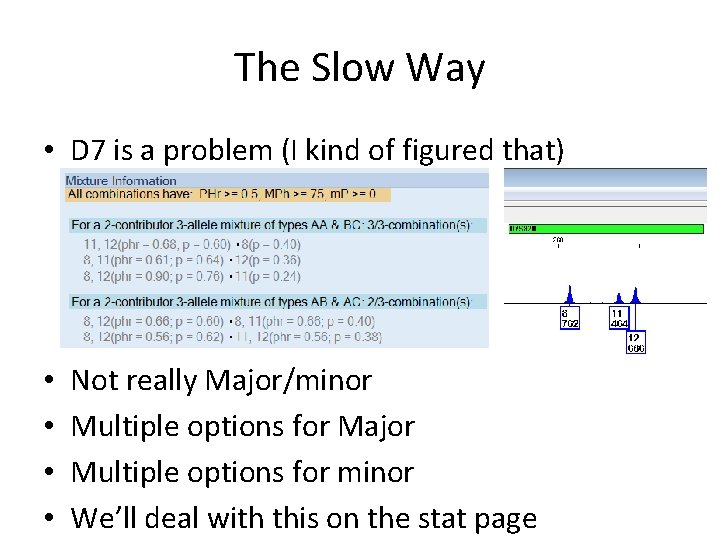
The Slow Way • D 7 is a problem (I kind of figured that) • • Not really Major/minor Multiple options for Major Multiple options for minor We’ll deal with this on the stat page

The Slow Way • At D 7 major is anything with a 12 except the 12, 12 – I’ll click in all three alleles into my major profile • At D 7 the minor is anything 8, 12 – I’ll click in all three alleles into my minor profile • On the stat page I have to pick the genotypes I want – both Major and minor are r. RMP here

The Slow Way • v. WA • 2 Alleles • Two options look OK • Two don’t • Probably not a 16, 16 and 17, 17 – 50/50 mixture • Probably not 16, 16 minor at 3% – That is 5 x less DNA than we’ve been seeing

The Slow Way • If we throw out the bad 50/50 and 97/3 • Major is 16, 17 and we are given either 16, 17 or 17, 17 for minor • But at smaller loci the minor is <300, so I’ll do 17, Any as minor at v. WA (Major is 16, 17)

The Slow Way • • FGA I know this locus doesn’t amp well I’ll punt on this minor If all other minor alleles were >300, maybe I’d at least do a 22, Any

The Slow Way • Let’s see how we did • We can match the Victim and Suspect references against what we just interpreted (partially deconvoluted)

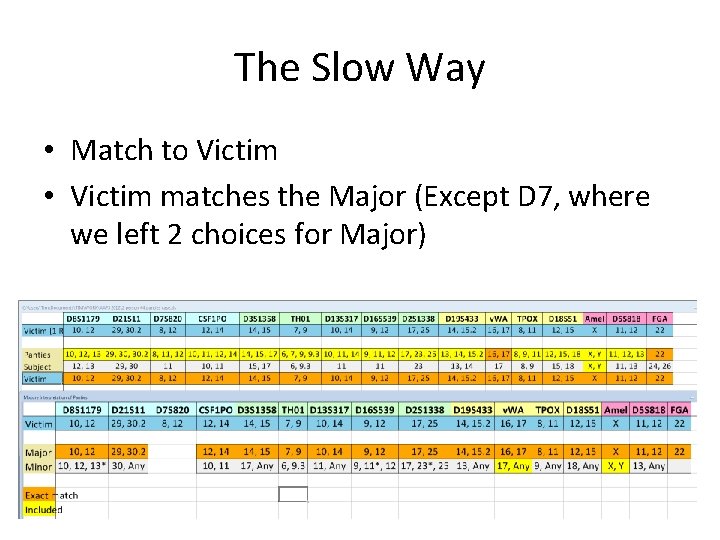
The Slow Way • Match to Victim • Victim matches the Major (Except D 7, where we left 2 choices for Major)

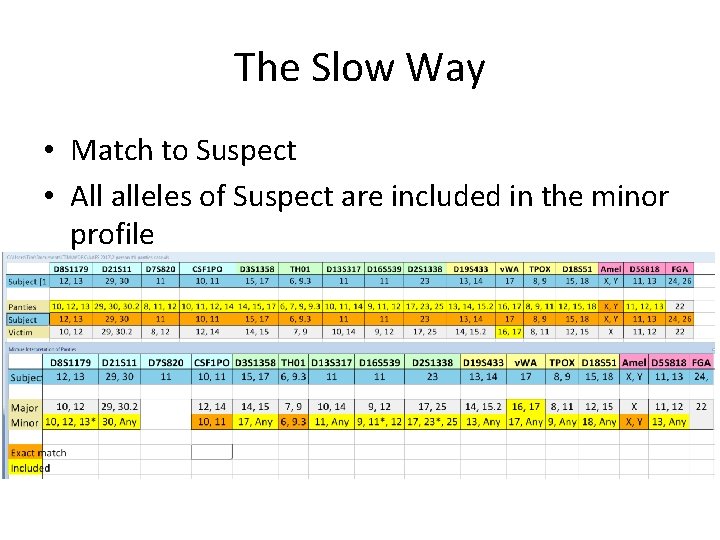
The Slow Way • Match to Suspect • All alleles of Suspect are included in the minor profile

The Slow Way • Match to Suspect (continued) • Some loci were Allele, Any but because the Suspect has that required allele, it lights up yellow • D 21 just needs 30

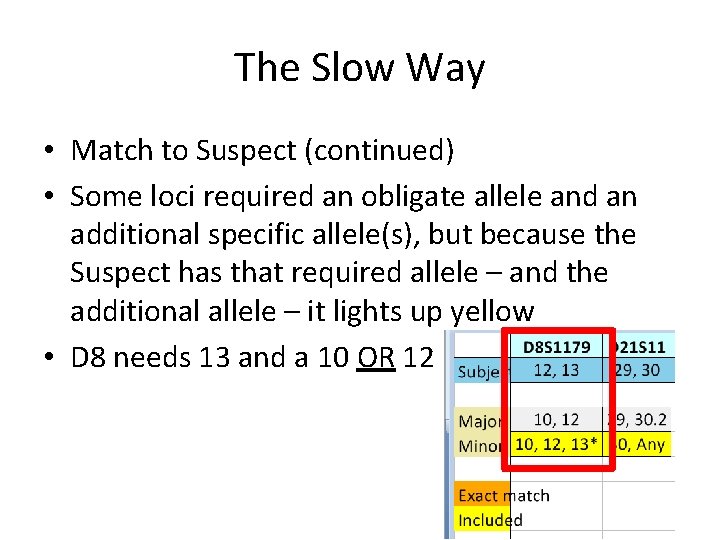
The Slow Way • Match to Suspect (continued) • Some loci required an obligate allele and an additional specific allele(s), but because the Suspect has that required allele – and the additional allele – it lights up yellow • D 8 needs 13 and a 10 OR 12

The Slow Way • Calculating the stat for the Major is quick • Just hit the “Mixture” button under “Frequency Calculations” ribbon • Yes, I know I said mixture and we came up with a single source profile • Not quite though – remember D 7? • The Major could be anything but a homozygote

The Slow Way • D 7 on the stat page with all three homozygote options • I realize you can’t read this, but there are 3 types calculated and summed up for the locus

The Slow Way • Final overall stat for the Major Contributor • 1 in 221 Quintillion

The Slow Way • Calculating the stat for the Minor is also quick • Just send it to the mixture stat page • This time, we really need the mixture as there are several loci where we have to consider more than one distinct genotype • Remember that obligate function?

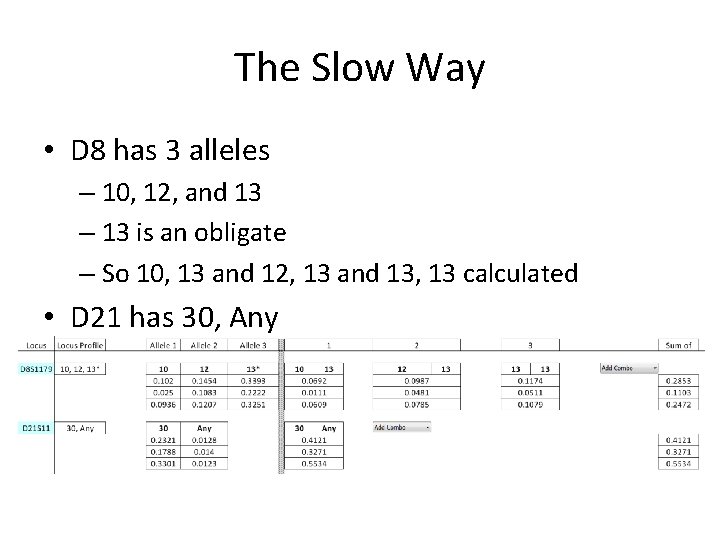
The Slow Way • D 8 has 3 alleles – 10, 12, and 13 – 13 is an obligate – So 10, 13 and 12, 13 and 13, 13 calculated • D 21 has 30, Any

The Slow Way • D 7 is also a problem here (no easy Major/minor) • But everything except 8, 12 is OK for the minor

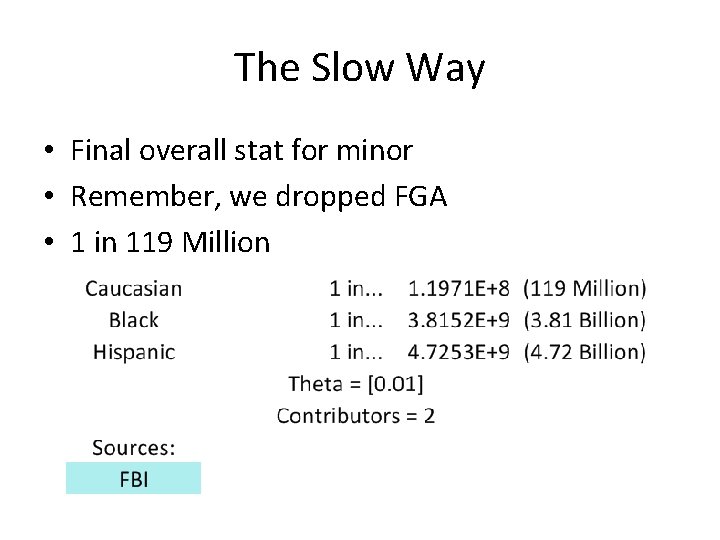
The Slow Way • Final overall stat for minor • Remember, we dropped FGA • 1 in 119 Million

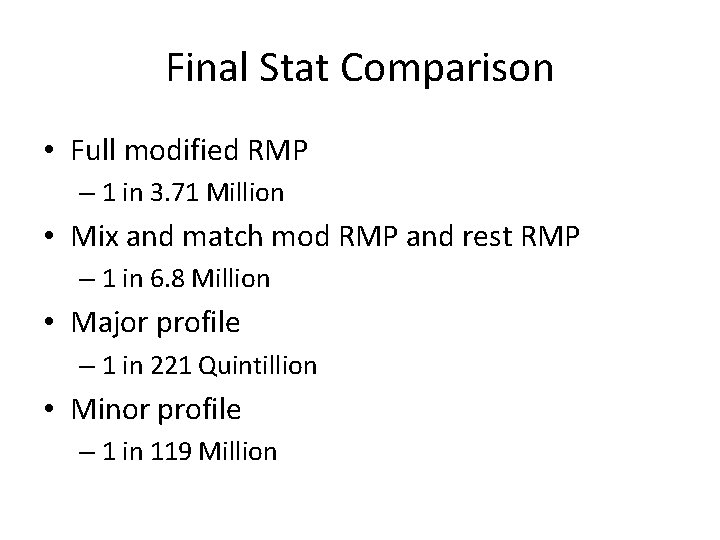
Final Stat Comparison • Full modified RMP – 1 in 3. 71 Million • Mix and match mod RMP and rest RMP – 1 in 6. 8 Million • Major profile – 1 in 221 Quintillion • Minor profile – 1 in 119 Million

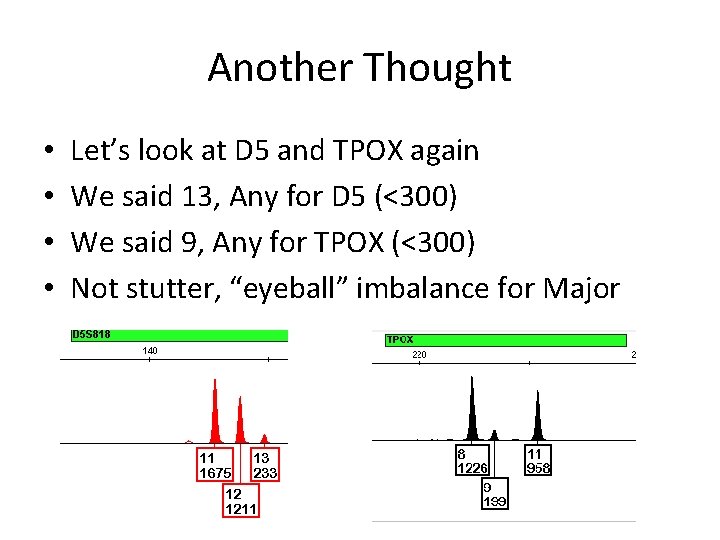
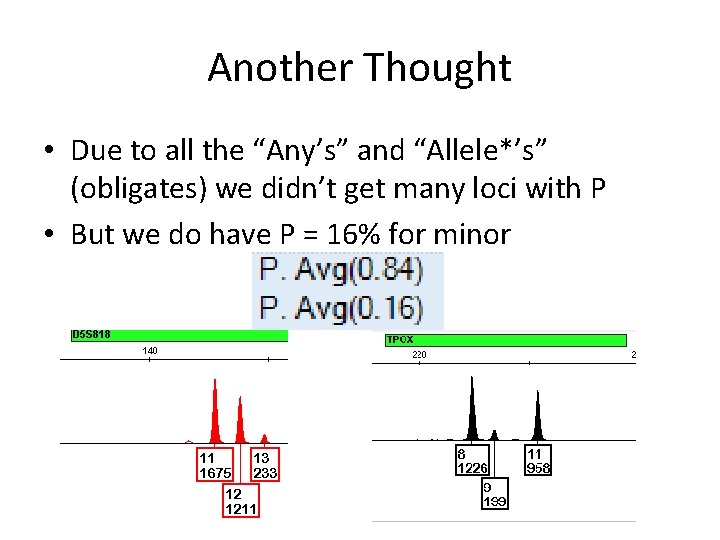
Another Thought • • Let’s look at D 5 and TPOX again We said 13, Any for D 5 (<300) We said 9, Any for TPOX (<300) Not stutter, “eyeball” imbalance for Major

Another Thought • Due to all the “Any’s” and “Allele*’s” (obligates) we didn’t get many loci with P • But we do have P = 16% for minor

Another Thought • Although the minor alleles are <300 at these two loci, they’re close enough to 300 that the probability of drop out may be small • Especially for D 5 at 233 rfu • Plus, we admit our 300 rfu threshold is on the “cautious” side – and we’re not happy about it • Some folks (Dr. Buckleton) would say the probability of drop out is low so “Any” is not the best approach to use – Continuous LR?

Another Thought • What does SWGDAM say? 3. 2. Application of Peak Height Thresholds to Allelic Peaks Amplification of low-level DNA samples may be subject to stochastic effects, where two alleles at a heterozygous locus exhibit considerably different peak heights (i. e. , peak height ratio generally <60%) or an allele fails to amplify to a detectable level (i. e. , allelic dropout). Stochastic effects within an amplification may affect one or more loci irrespective of allele size. Such low-level samples exhibit peak heights within a given range which is dependent on quantitation system, amplification kit and detection instrumentation. A threshold value can be applied to alert the DNA analyst that all of the DNA typing information may not have been detected for a given sample. This threshold, referred to as a stochastic threshold, is defined as the value above which it is reasonable to assume that allelic dropout has not occurred within a single-source sample. The application of a stochastic threshold to the interpretation of mixtures should take into account the additive effects of potential allele sharing.

Another Thought • To me that means we don’t automatically have to assume drop out when in the “Danger Zone” • Especially when I have a great tool to investigate “the additive effects of potential allele sharing” • So if my math (PHR and P) shows me I see two alleles of the minor, but one is shared by the Major, I can use a restricted RMP (SS maybe? )

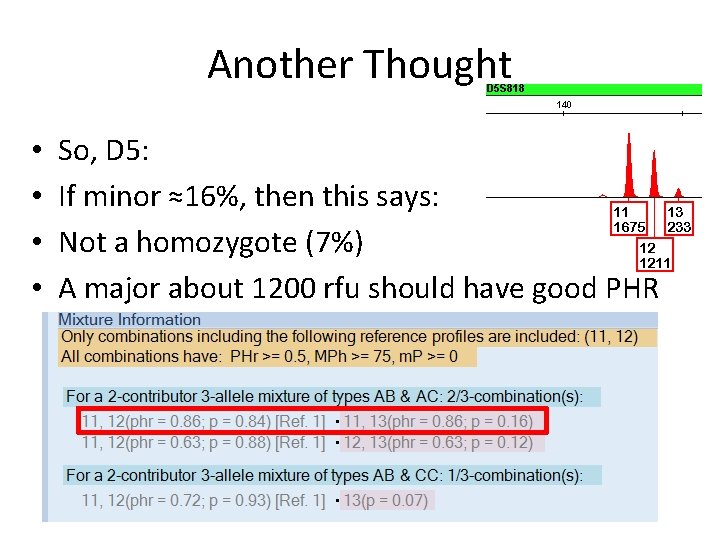
Another Thought • • So, D 5: If minor ≈16%, then this says: Not a homozygote (7%) A major about 1200 rfu should have good PHR

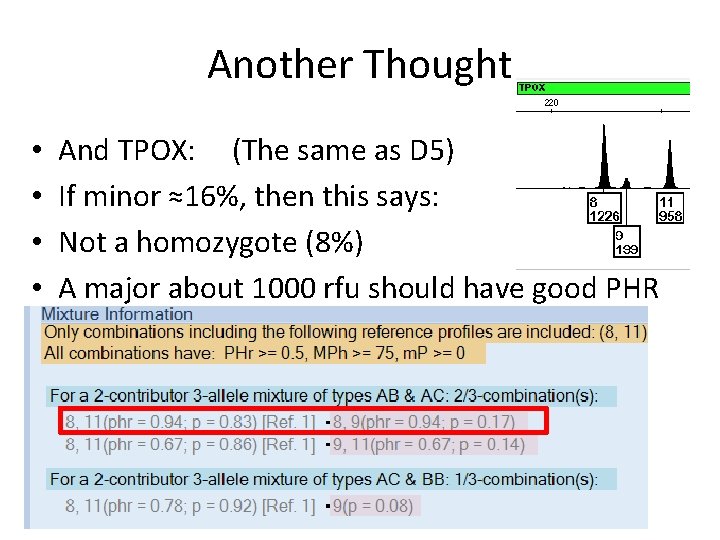
Another Thought • • And TPOX: (The same as D 5) If minor ≈16%, then this says: Not a homozygote (8%) A major about 1000 rfu should have good PHR

Another Thought • Why would ½ the DNA of the minor just disappear? The “real” chance of drop out is probably pretty low • If you consider masking, you just found the other ½ of the minor DNA

What about a LR? • The LR assumes contributors • We set up 2 competing hypotheses • It is essentially one RMP divided by a different RMP (sort of) • But how do you choose the hypotheses?

What about LR • Hp says “Victim and this Suspect!” • Would Hd say that “because the panties were found under the bush it’s 2 unknowns? ” • They might, but they shouldn’t – If V + S alleles are present, the LR is usually impressive enough if Hd is V + U – If V + S alleles are present, and the Hd is U + U, the LR usually becomes crazy

What about LR • Can we use LR here? • All alleles of both people are present • But… – When we did the RMP, we allowed for drop out due to low level alleles – If we must account for potential drop out in the RMP, why would we not do so for the LR?

What about LR • Remember, the Hd says “It may be the Victim, but it’s not my client” • Furthermore, Hd may say “Not only is it not my client, but you may be missing alleles from the REAL bad guy, so your LR calculation is not fair to my client”

What about LR • So, we have to consider that there may be drop out. • At v. WA we didn’t detect any minor alleles at all, defense says we need to be concerned about 2 alleles dropping out

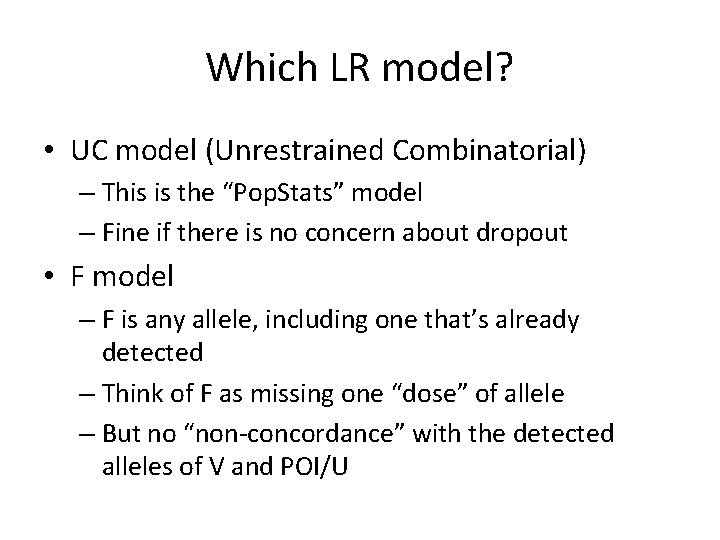
Which LR model? • UC model (Unrestrained Combinatorial) – This is the “Pop. Stats” model – Fine if there is no concern about dropout • F model – F is any allele, including one that’s already detected – Think of F as missing one “dose” of allele – But no “non-concordance” with the detected alleles of V and POI/U

Which LR model? • Q model – Doesn’t directly deal with drop out, but it does work Θ back in for homozygotes – Allows for dealing with distinct genotypes in a LR

Which LR model? • The non-concordance model – “The D model” – We can allow for multiple stochastic events – Need to combine with F or Q model for concordant alleles – Can restrict based on phr models – Can get pretty messy to calculate by hand

D Model • Can be used with non-concordance – Ex: Locus has 11, 12, 13 alleles – V is 11, 12 – POI is 13, 14 – a non-concordance • This example we’ve been working with doesn’t have non-concordance (except for FGA), but Hd says “The real guy may have dropped out” so we need D model if we’re to do LR

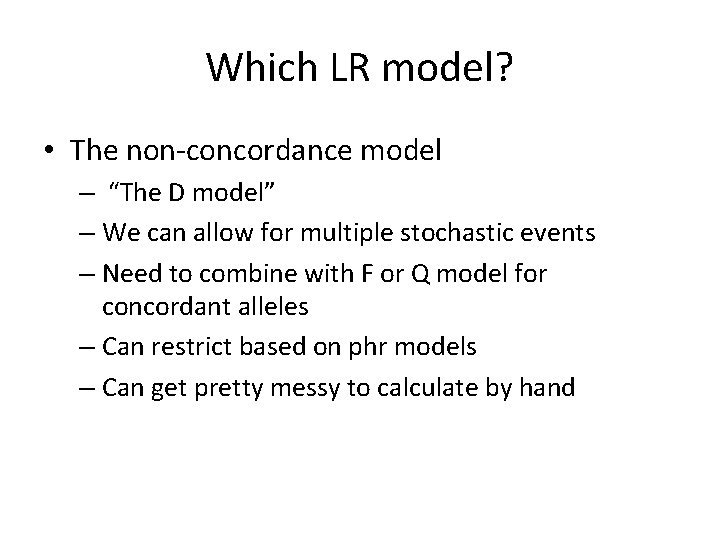
D Model • • v. WA V is 16, 17 POI is 17, 17 Hd says “Yea, but it’s our theory that both alleles of the REAL bad guy have dropped out”

D Model • We need to introduce 2 terms – Drop • The probability that an allele dropped out • You have to determine this, probably related to the height of the minor alleles that you do see elsewhere – Not Drop • The probability that an allele did not drop out • (This one is pretty easy, if you see the allele, it didn’t drop out. )

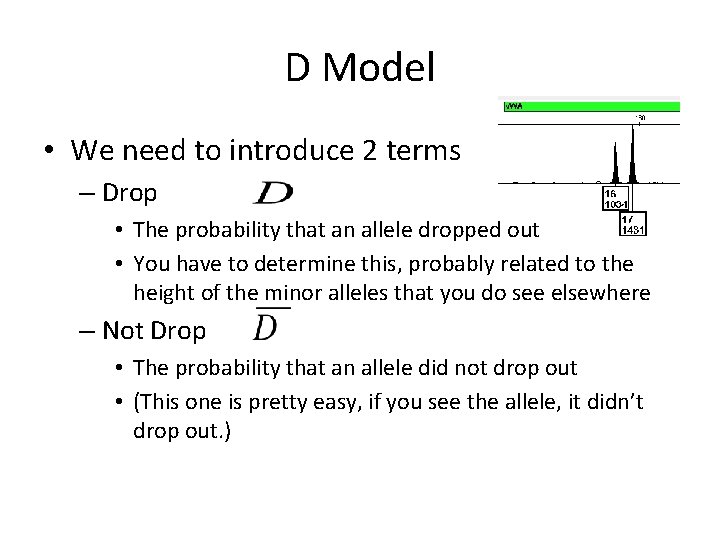
D Model • Hp says both are included – Must account for 16, 17 and 17, 17 – But since the 17 is accounted for by V

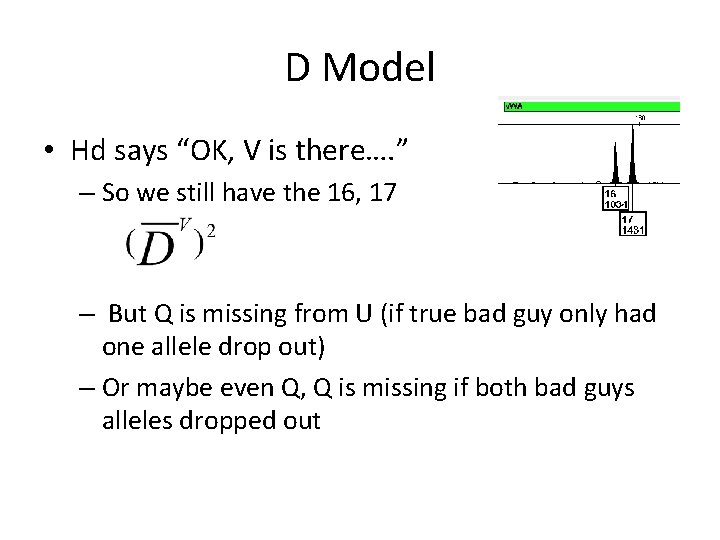
D Model • Hd says “OK, V is there…. ” – So we still have the 16, 17 – But Q is missing from U (if true bad guy only had one allele drop out) – Or maybe even Q, Q is missing if both bad guys alleles dropped out

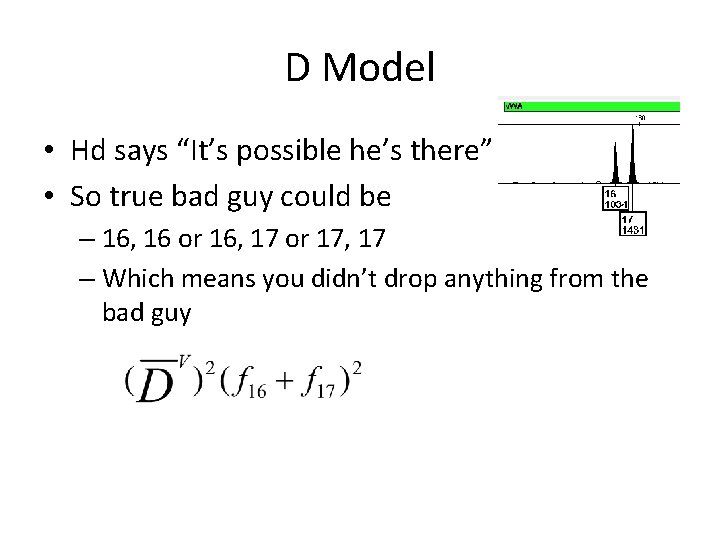
D Model • Hd says “It’s possible he’s there” • So true bad guy could be – 16, 16 or 16, 17 or 17, 17 – Which means you didn’t drop anything from the bad guy

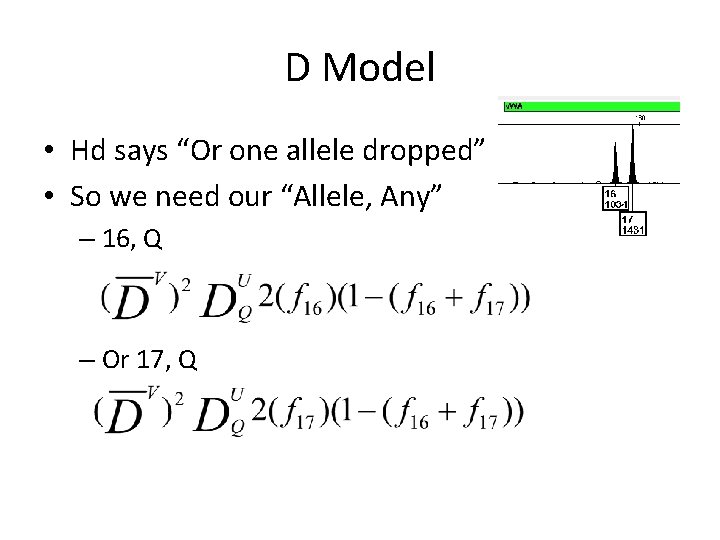
D Model • Hd says “Or one allele dropped” • So we need our “Allele, Any” – 16, Q – Or 17, Q

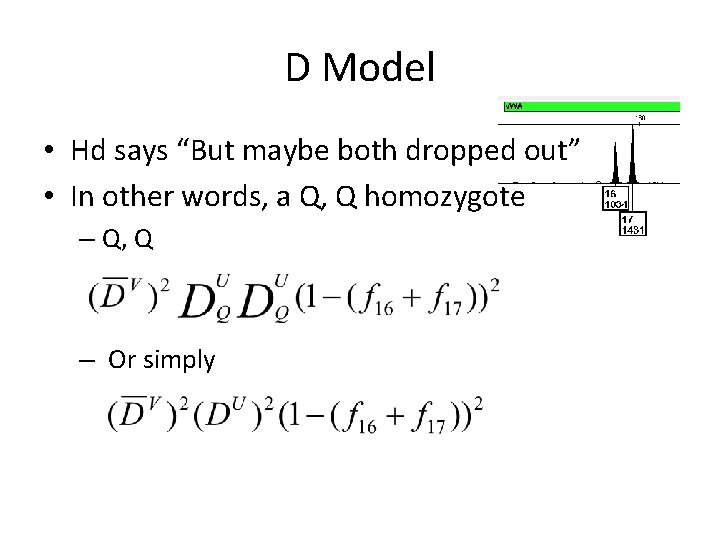
D Model • Hd says “But maybe both dropped out” • In other words, a Q, Q homozygote – Q, Q – Or simply

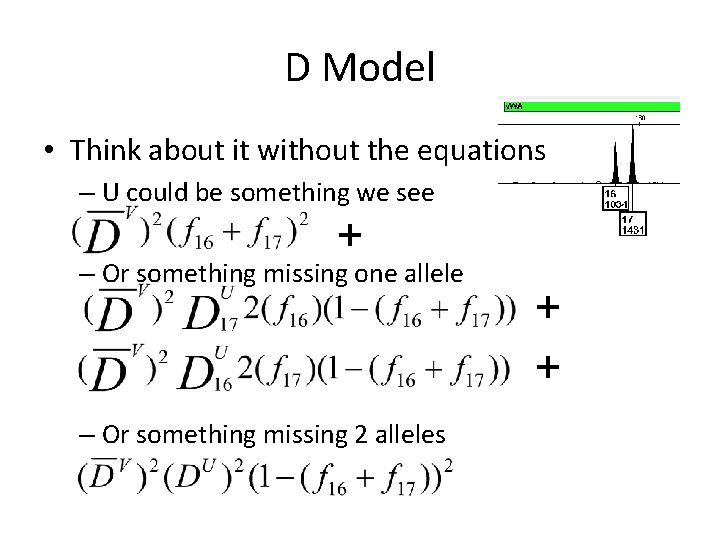
D Model • Think about it without the equations – U could be something we see + – Or something missing one allele – Or something missing 2 alleles + +

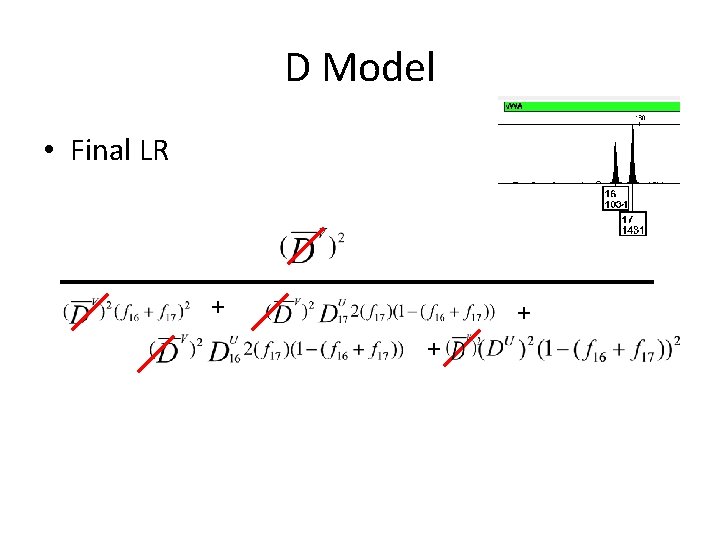
D Model • Final LR + + +

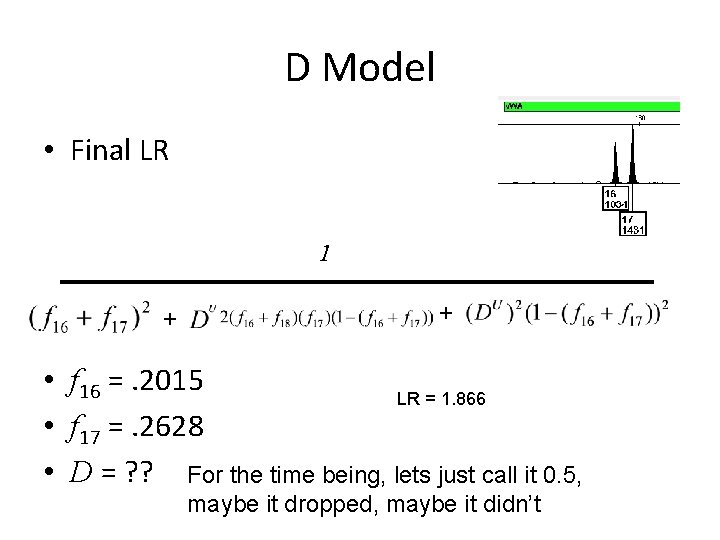
D Model • Final LR 1 + + • f 16 =. 2015 LR = 1. 866 • f 17 =. 2628 • D = ? ? For the time being, lets just call it 0. 5, maybe it dropped, maybe it didn’t

The Egram