Where to get Perl www perl com www











- Slides: 13

Where to get Perl ? www. perl. com / www. perl. org n Unix/Linux/Solaris n n n Interpreter (called “perl”) comes pre-installed usually Get source from www. perl. com Win 32 n Get source distribution from www. perl. com n n Get binary from www. activestate. com n n No need to compile, runs pretty well Macintosh n n Have to compile www. macperl. com Perl IDE (integrated development environment) n Open Perl IDE (Jurgen Guntherodt) n n It’s mainly an editor. Active. State Komodo 3. 0 (IDE for Perl, Python, Tcl, …) n $29. 95 for a student license Visit the class web site for the links!

Running Perl: Windows n Open a text editor such as Notepad 1. 2. n write your perl code save your code with an extension “. pl” Open MS-DOS/Command window 1. 2. 3. 4. 5. go to your folder in which your code is saved make a shortcut to the command window (enter “cmd” to make the shortcut) right click on the icon and go to properties clear the “start in” field Run your code as “perl <filename>” or 1. 2. go to Start → run → “cmd” , … run your code as “perl <filename>”

Programming and Perl for Bioinformatics Part II

Basic Data Types n Perl has three basic data types: n scalar n array (list) n associative array (hash)

Arrays n An array (list) is an ordered group of scalar values. n ‘@’ is used to refer to the entire array Example: n n n (1, 2, 3) # Array of three values 1, 2, and 3 ("one", "two", "three") # Array of 3 values "one", "two", "three" @names = ("mary", "tom", "mark", "john", "jane"); #? “tom” $names [1] ; Extract 2 nd item from @names n @names [1. . 4]; Extract the sublist from @names

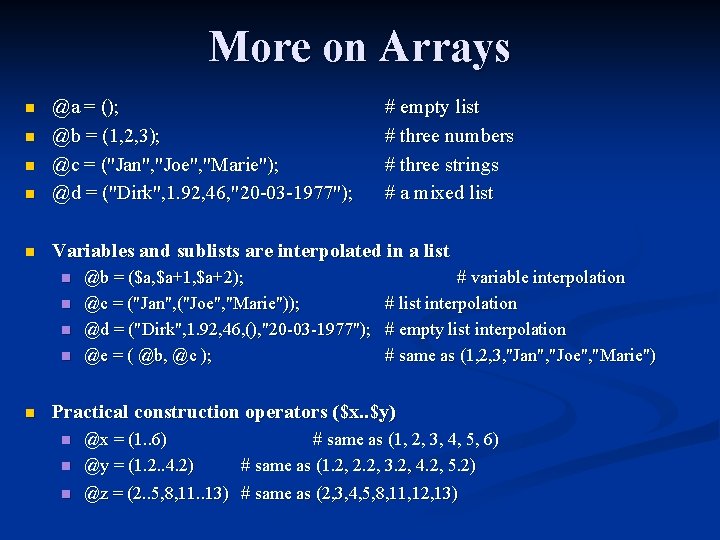
More on Arrays n @a = (); @b = (1, 2, 3); @c = ("Jan", "Joe", "Marie"); @d = ("Dirk", 1. 92, 46, "20 -03 -1977"); n Variables and sublists are interpolated in a list n n n n # empty list # three numbers # three strings # a mixed list @b = ($a, $a+1, $a+2); # variable interpolation @c = ("Jan", ("Joe", "Marie")); # list interpolation @d = ("Dirk", 1. 92, 46, (), "20 -03 -1977"); # empty list interpolation @e = ( @b, @c ); # same as (1, 2, 3, "Jan", "Joe", "Marie") Practical construction operators ($x. . $y) n @x = (1. . 6) @y = (1. 2. . 4. 2) n @z = (2. . 5, 8, 11. . 13) # same as (2, 3, 4, 5, 8, 11, 12, 13) n # same as (1, 2, 3, 4, 5, 6) # same as (1. 2, 2. 2, 3. 2, 4. 2, 5. 2)

Array Manipulations reverse Reverses the order of array elements @a = (1, 2, 3); @b = reverse @a; # @b = (3, 2, 1); split Splits a string into a list/array $line = "John Smith 28"; ($first, $last, $age) = split /s/, $line; $DNA = "ACGTTTGA"; @DNA = split ('', $DNA); join Joins a list/array into a string $gene = join "", ($exon 1, $exon 3); $name = join "-", ("Zhong", "Hui"); scalar Returns the number of elements in @array scalar @array; sort Return sorted elements sort { $a <=> $b } @not_sorted sort { $a cmp $b } @not_sorted # numerical sort # ASCII-betical sort

Exercise #Determine freq of nucleotides $dna ="ga. Tt. ACata. CACTgttca"; ?

Filehandles File I/O (input/output): reading from/writing to files n Files represented in Perl by a filehandle variable (for clarity, usu. written as a bare word in UPPERCASE) n Open a file on a filehandle using the open function n n for reading (input): open INFILE, “< datafile. txt”; or open (INFILE, “< datafile. txt”); for writing (output), overwriting the file: open OUTFILE, “> output”; for appending to the end of the file: open OUTFILE, “>> output”; Close a file on a filehandle n Close (OUTFILE);

Special Filehandles Special “files” that are always “open” n STDIN (standard input) n input from command window read only n STDOUT (standard output) n output to command window write only print STDOUT “Have fun with Perl!n”; or just print “Have fun with Perl!n”;

Input from Filehandles “Angle Bracket” input operator reads one line of input (up to newline/carriage return) from STDIN: n n print $line chomp print n "Enter name of protein: "; = <STDIN>; $line; # removes n from end of $line “n. You entered $line. n”; from a file: open (INPUT, “aminos. txt”); $amino 1 = <INPUT>; $amino 2 = <INPUT>; chomp ($amino 1, $amino 2);

sequences. fasta >gi|145536|gb|L 04574. 1|Escherichia coli DNA polymerase III chi subunit (hol. C) gene, complete cds TAACGGCGAAGAGTAATTGCGTCAGGCAAGGCTGTTATTGCCGGATGCGGCGTGAACG CCTTATCCGACC TACACAGCACTGAACTCGTAGGCCTGATAAGACACAACAGCGTCGCATCAGGCGCTGC GGTGTATACCTG ATGCGTATTTAAATCCACCACAAGAAGCCCCATTTATGAAAAACGCGACGTTCTACCTTC TGGACAATGA CACCACCGTCGATGGCTTAAGCGCCGTTGAGCAACTGGTGTGTGAAATTGCCGCAGAAC GTTGGCGCAGC GGTAAGCGCGTGCTCATCGCCTGTGAAGATGAAAAGCAGGCTTACCGGCTGGATGAAG CCCTGTGGGCGC GTCCGGCAGAAAGCTTTGTTCCGCATAATTTAGCGGGAGAAGGACCGCGCGGCGGTGC ACCGGTGGAGAT CGCCTGGCCGCAAAAGCGTAGCAGCAGCCGGCGCGATATATTGATTAGTCTGCGAACA AGCTTTGCAGAT TTTGCCACCGCTTTCACAGAAGTGGTAGACTTCGTTCCTTATGAAGATTCTCTGAAACAA CTGGCGCGCG AACGCTATAAAGCCTACCGCGTGGCTGGTTTCAACCTGAATACGGCAACCTGGAAATAA TGGAAAAGACA TATAACCCACAAGATATCGAACAGCCGCTTTACGAGCACTGGGAAAAGCAGGGCTACTT TAAGCCTAATG

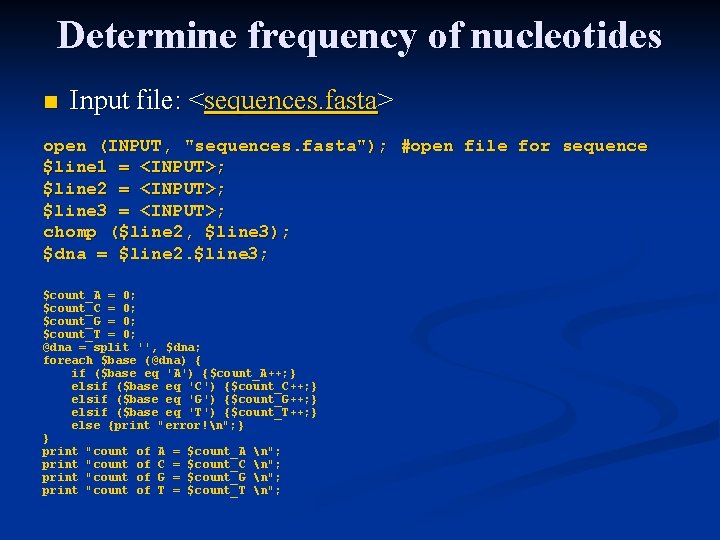
Determine frequency of nucleotides n Input file: <sequences. fasta> open (INPUT, "sequences. fasta"); #open file for sequence $line 1 = <INPUT>; $line 2 = <INPUT>; $line 3 = <INPUT>; chomp ($line 2, $line 3); $dna = $line 2. $line 3; $count_A = 0; $count_C = 0; $count_G = 0; $count_T = 0; @dna = split '', $dna; foreach $base (@dna) { if ($base eq 'A') {$count_A++; } elsif ($base eq 'C') {$count_C++; } elsif ($base eq 'G') {$count_G++; } elsif ($base eq 'T') {$count_T++; } else {print "error!n"; } } print "count of A = $count_A n"; print "count of C = $count_C n"; print "count of G = $count_G n"; print "count of T = $count_T n";