What happened to computing 1930 80 is now











- Slides: 56

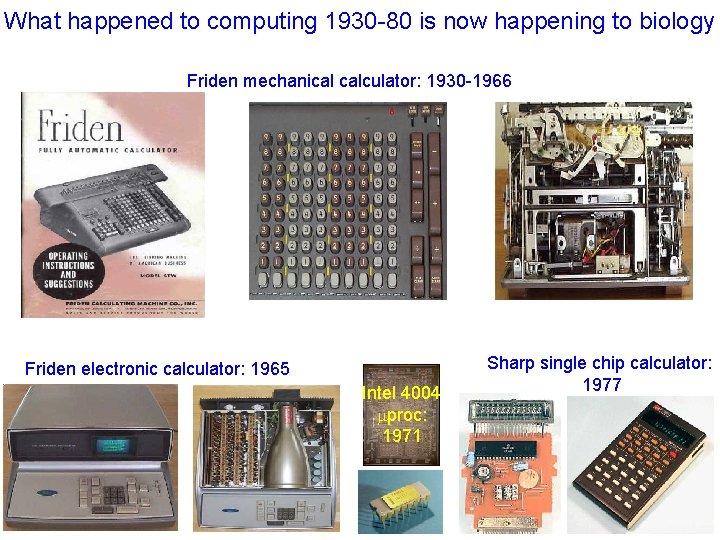
What happened to computing 1930 -80 is now happening to biology Friden mechanical calculator: 1930 -1966 Friden electronic calculator: 1965 Intel 4004 mproc: 1971 Sharp single chip calculator: 1977

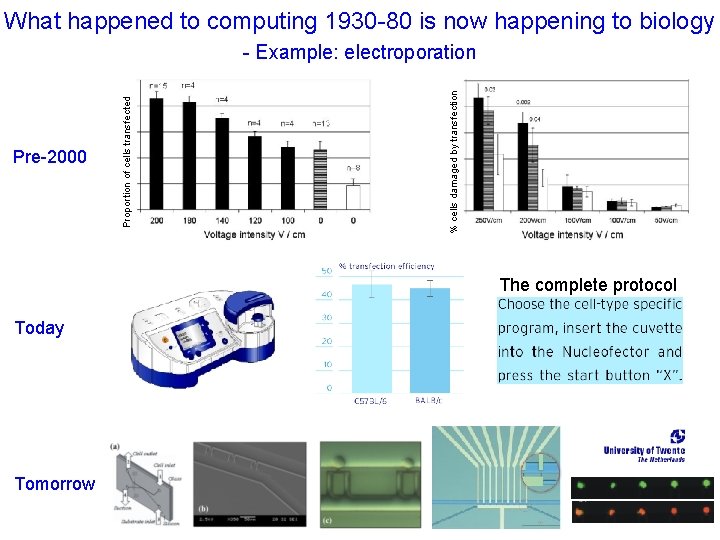
What happened to computing 1930 -80 is now happening to biology % cells damaged by transfection Pre-2000 Proportion of cells transfected - Example: electroporation The complete protocol Today Tomorrow

Another example: automated parallel mini-preps

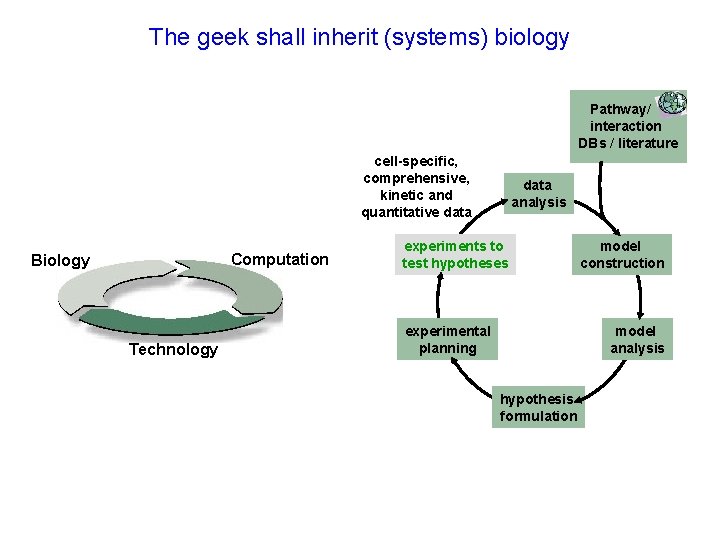
The geek shall inherit (systems) biology Pathway/ interaction DBs / literature cell-specific, comprehensive, kinetic and quantitative data Computation Biology Technology data analysis experiments to test hypotheses experimental planning model construction model analysis hypothesis formulation

2 complementary approaches in systems biology: Top-down, global, systematic Bottom-up, local dynamics • • Focus on nonlinear interactions • Study irreducible systems • Analyzing emergent (difficult-topredict, nonlinear) properties • Hypothesis and model-driven • Discovery science via high throughput ‘omic technologies Screening - multi-target/multi-component drugs - multi-parameter disease signatures • Observation & data-driven global climate local weather

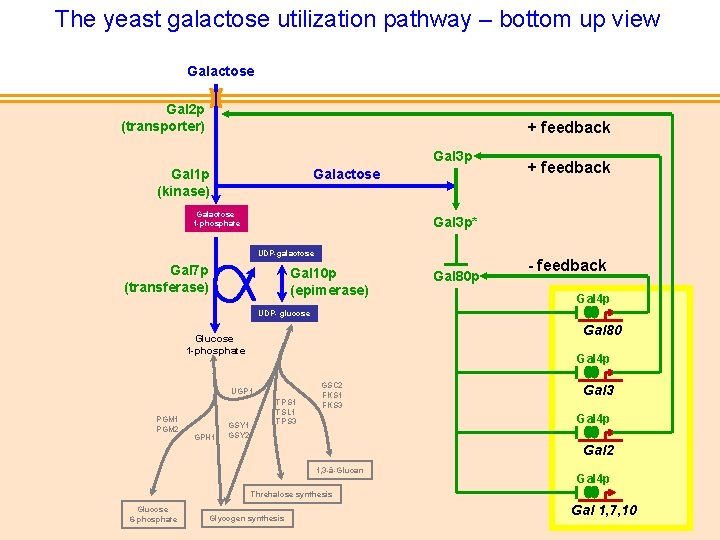
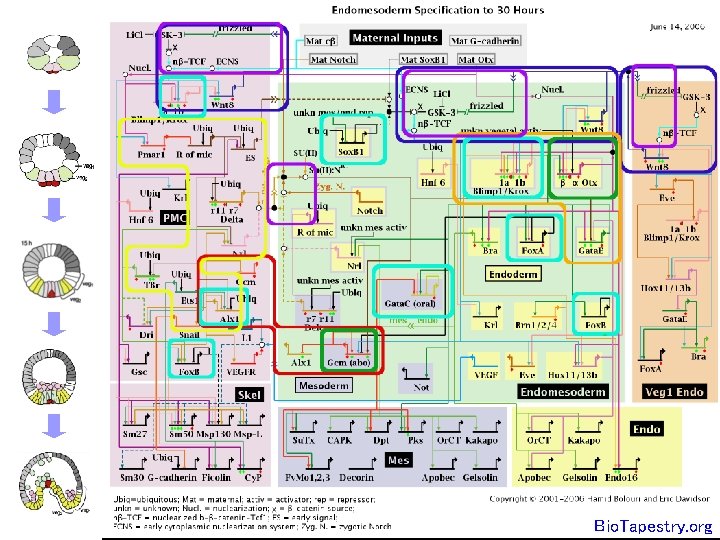
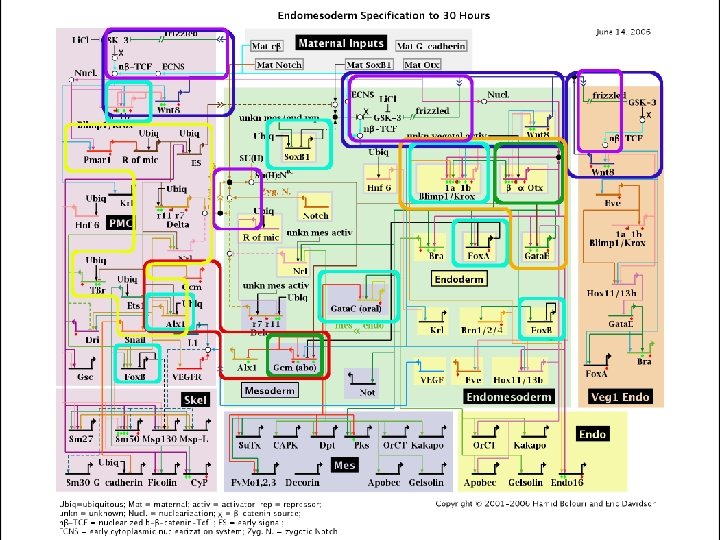
The yeast galactose utilization pathway – bottom up view Galactose Gal 2 p (transporter) + feedback Gal 3 p Gal 1 p (kinase) Galactose 1 -phosphate + feedback Gal 3 p* UDP-galactose Gal 7 p (transferase) Gal 10 p (epimerase) Gal 80 p - feedback Gal 4 p UDP- glucose Gal 80 de Atauri et al Biochem J. 2005 Glucose 1 -phosphate Gal 4 p UGP 1 PGM 2 GPH 1 GSY 2 TPS 1 TSL 1 TPS 3 GSC 2 FKS 1 FKS 3 Gal 4 p Gal 2 1, 3 -â-Glucan Gal 4 p Threhalose synthesis Glucose 6 -phosphate Glycogen synthesis Gal 1, 7, 10

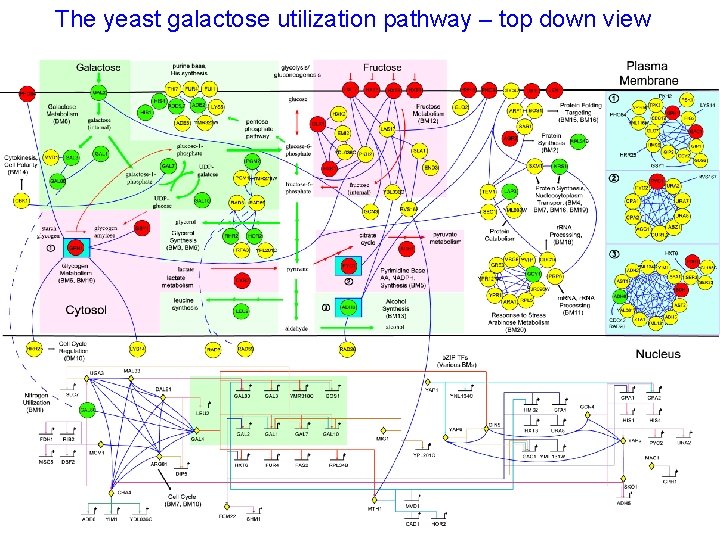
The yeast galactose utilization pathway – top down view

How do we make sense of this?

A network graph viewed in Cytoscape; what does it do?

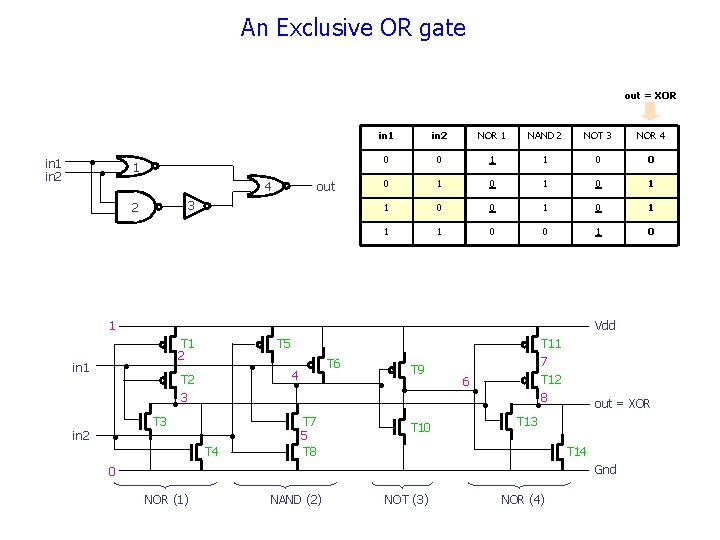
An Exclusive OR gate out = XOR • in 1 in 2 1 • 4 out 3 2 in 1 in 2 NOR 1 NAND 2 NOT 3 NOR 4 0 0 1 1 0 0 0 1 0 1 1 0 0 1 0 1 T 1 2 • in 1 in 2 Vdd • T 5 . • T 2 3 T 4 • T 11 T 6 4 • • • T 7 5 T 8 T 9 T 10 6 • • 7 T 12 8 • out = XOR T 13 T 14 Gnd 0 NOR (1) NAND (2) NOT (3) NOR (4)

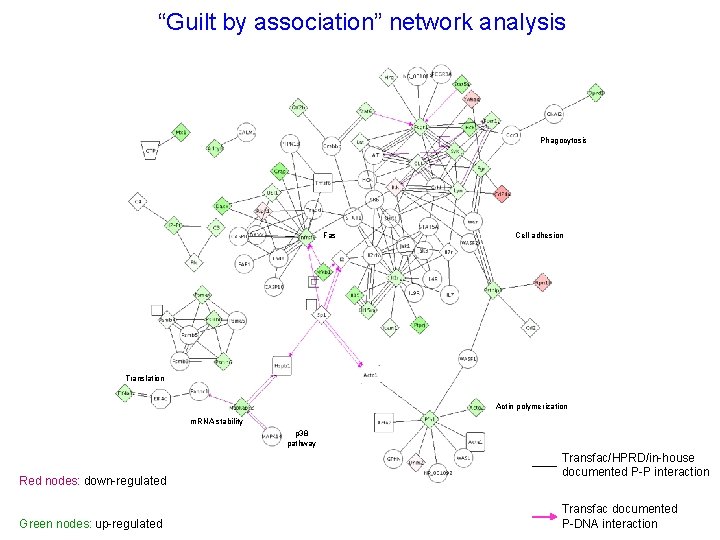
“Guilt by association” network analysis Phagocytosis Fas Cell adhesion Translation Actin polymerization m. RNA stability p 38 pathway Red nodes: down-regulated Green nodes: up-regulated Transfac/HPRD/in-house documented P-P interaction Transfac documented P-DNA interaction

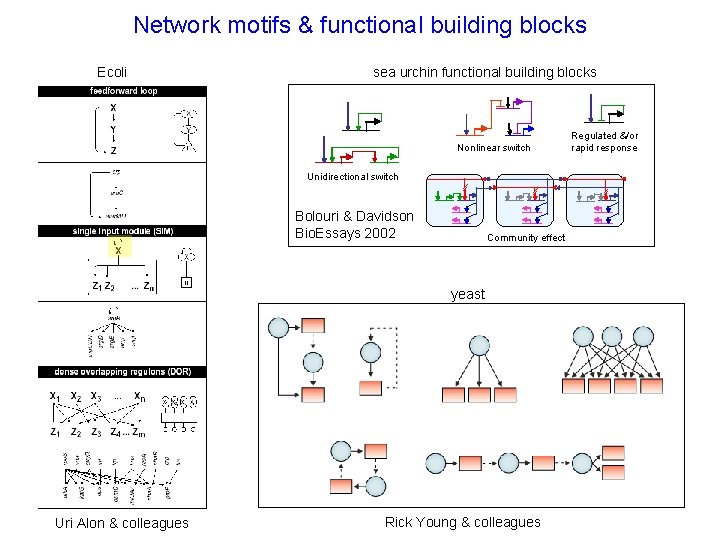
Network motifs & functional building blocks Ecoli sea urchin functional building blocks Nonlinear switch Unidirectional switch Bolouri & Davidson Bio. Essays 2002 Community effect yeast Uri Alon & colleagues Rick Young & colleagues Regulated &/or rapid response

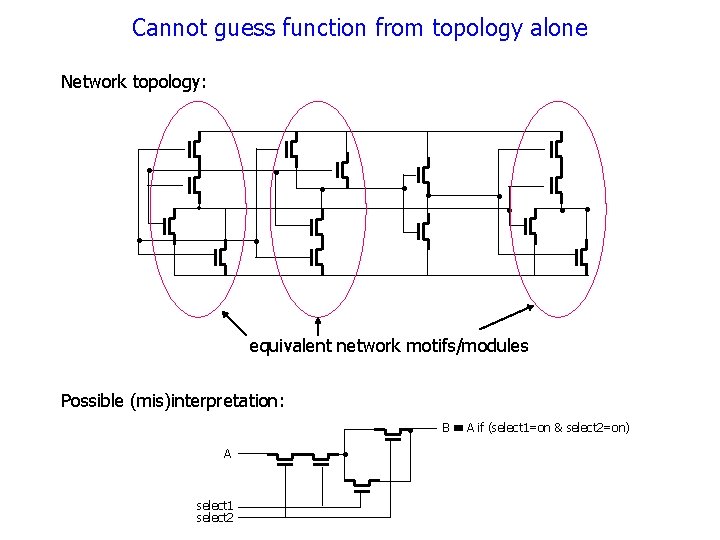
Cannot guess function from topology alone Network topology: 1 • in 1 in 2 T 1 2 • . T 5 • T 2 T 3 T 4 • T 11 T 6 • • • 7 T 12 T 9 • • T 10 5 T 8 • • T 13 T 14 0 equivalent network motifs/modules select 1 select 2 • • T 13 7 T 12 A T 11 Possible (mis)interpretation: B A if (select 1=on & select 2=on)

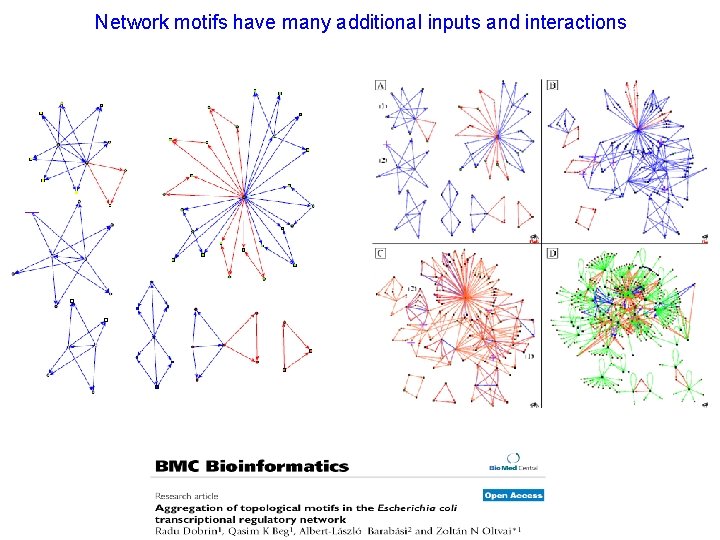
Network motifs have many additional inputs and interactions

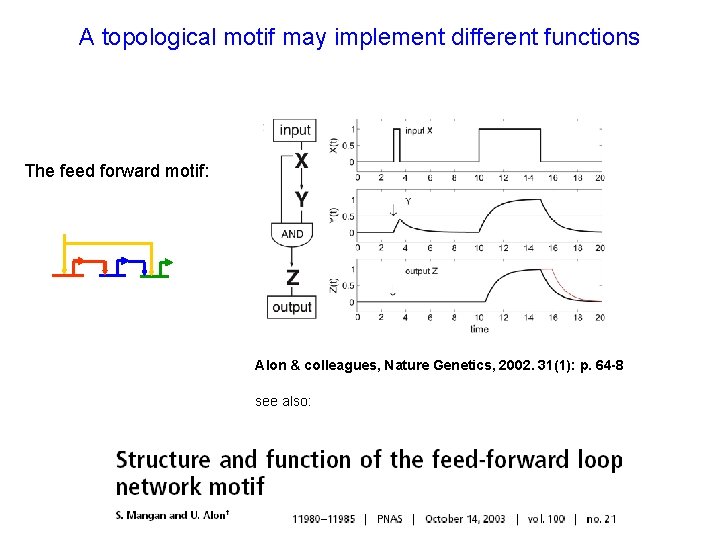
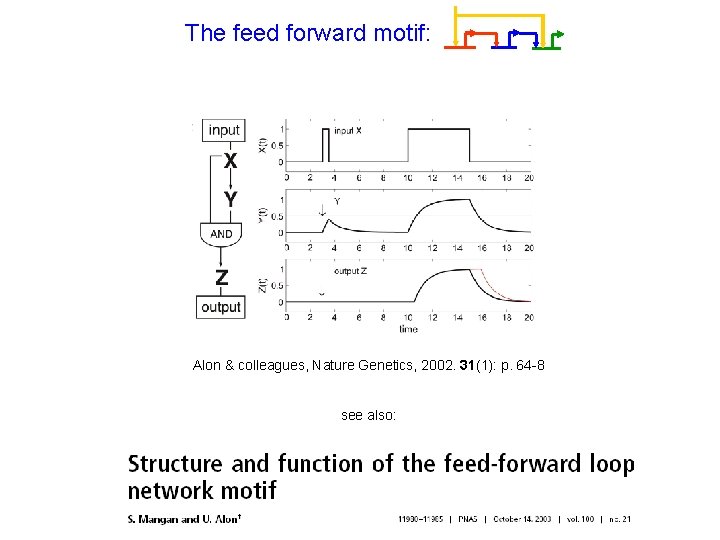
A topological motif may implement different functions The feed forward motif: Alon & colleagues, Nature Genetics, 2002. 31(1): p. 64 -8 see also:

A coherent feedforward motif acting as a gradient sensor

Need a functional abstraction hierarchy

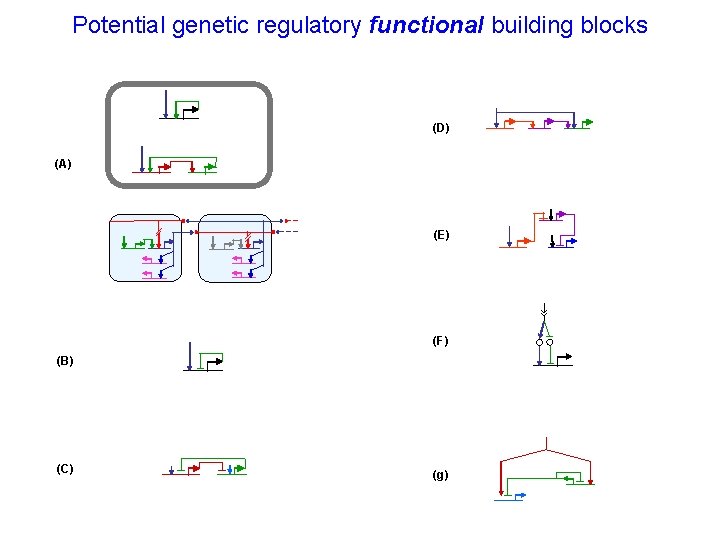
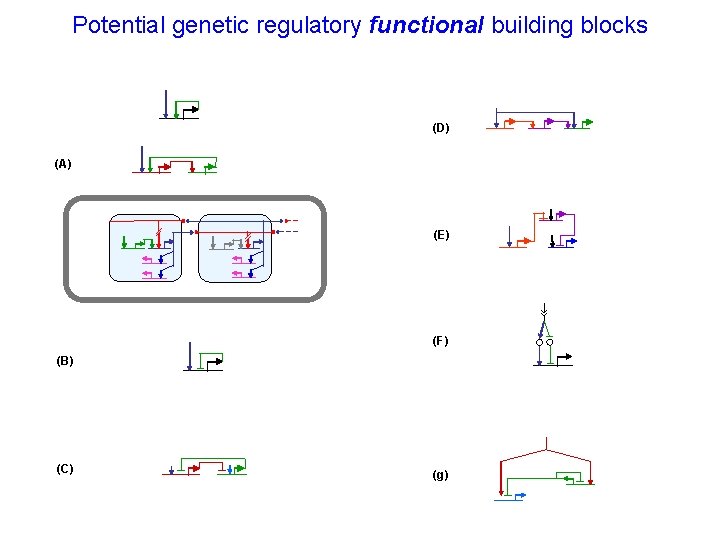
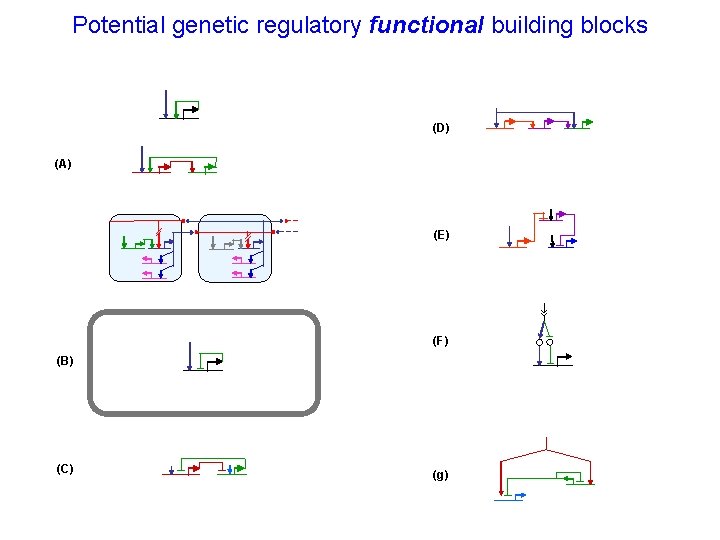
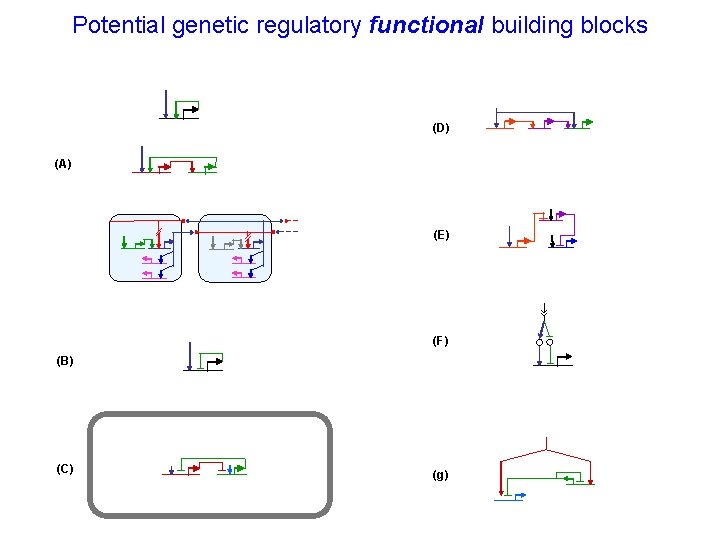
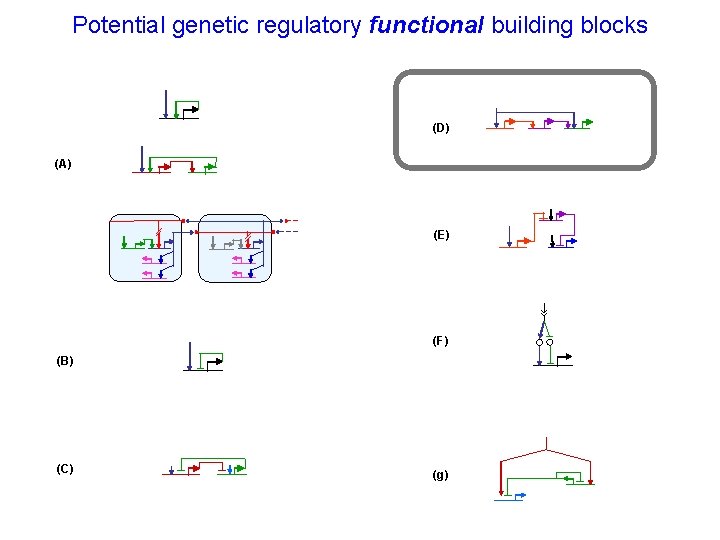
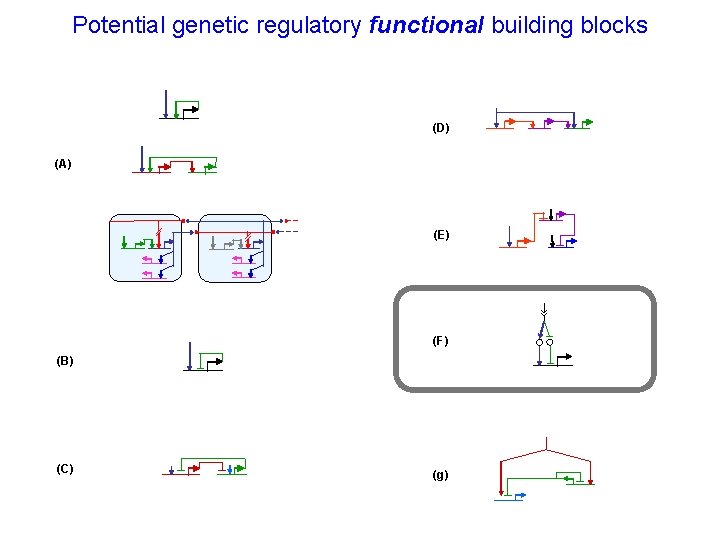
Potential genetic regulatory functional building blocks (D) (A) (E) (F) (B) (C) (g)

Bio. Tapestry. org


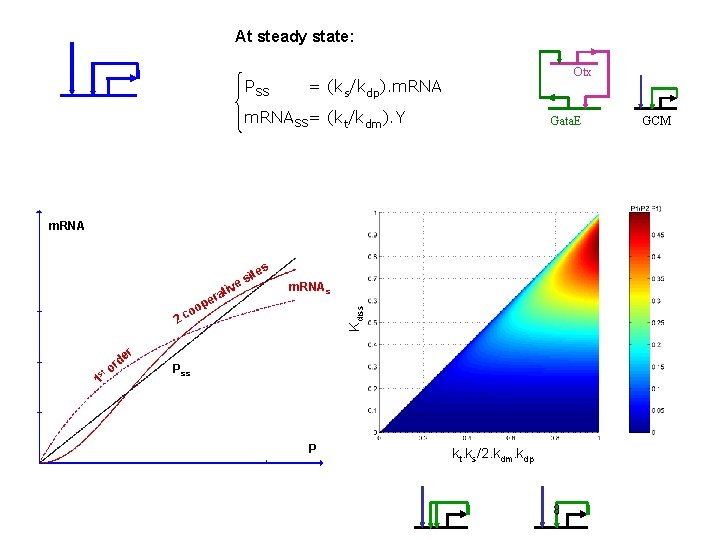
At steady state: PSS Otx = (ks/kdp). m. RNASS= (kt/kdm). Y Gata. E m. RNA es iv it es t era m. RNAss Kdiss p oo c 2 r st 1 de or Pss P kt. ks/2. kdm. kdp GCM

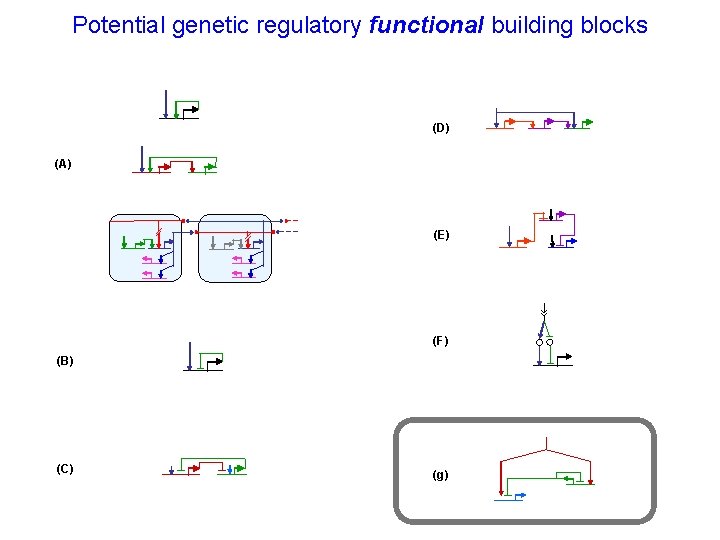
Potential genetic regulatory functional building blocks (D) (A) (E) (F) (B) (C) (g)


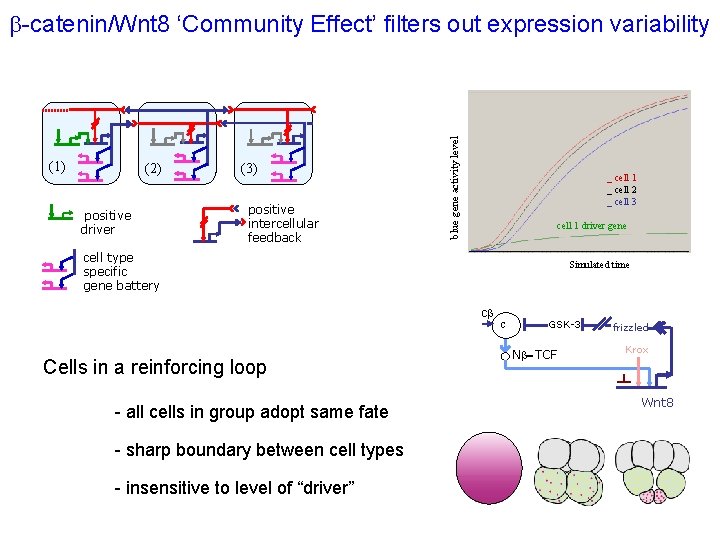
(1) (2) positive driver (3) positive intercellular feedback blue gene activity level b-catenin/Wnt 8 ‘Community Effect’ filters out expression variability _ cell 1 _ cell 2 _ cell 3 cell 1 driver gene cell type specific gene battery Simulated time cb Cells in a reinforcing loop - all cells in group adopt same fate - sharp boundary between cell types - insensitive to level of “driver” c GSK-3 Nb-TCF frizzled Krox Wnt 8

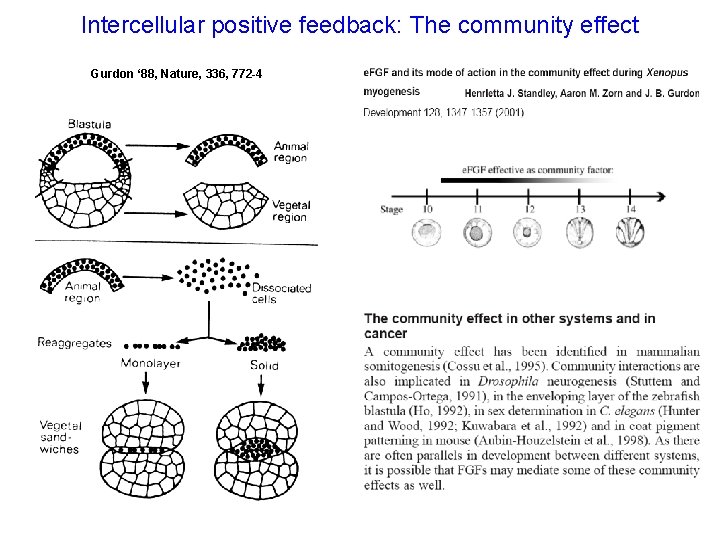
Intercellular positive feedback: The community effect Gurdon ‘ 88, Nature, 336, 772 -4

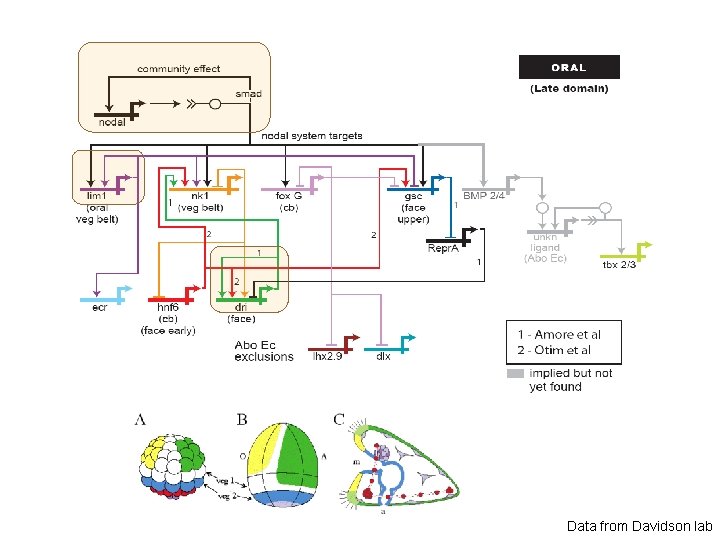
Data from Davidson lab

Potential genetic regulatory functional building blocks (D) (A) (E) (F) (B) (C) (g)

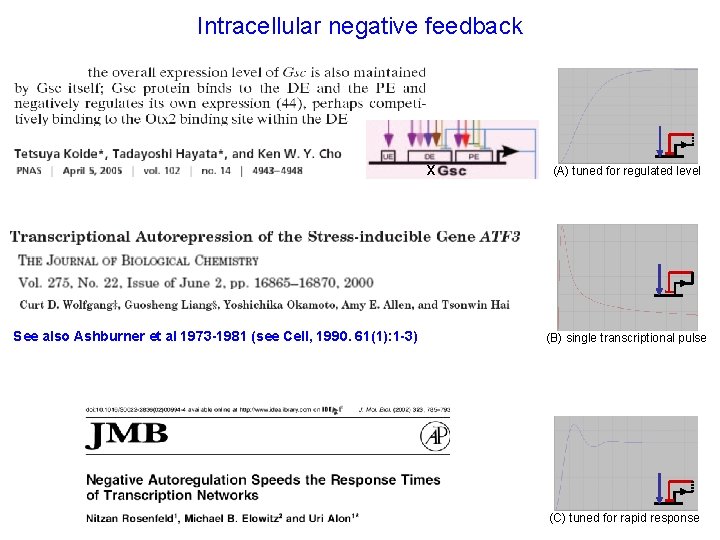
Intracellular negative feedback x See also Ashburner et al 1973 -1981 (see Cell, 1990. 61(1): 1 -3) (A) tuned for regulated level (B) single transcriptional pulse (C) tuned for rapid response

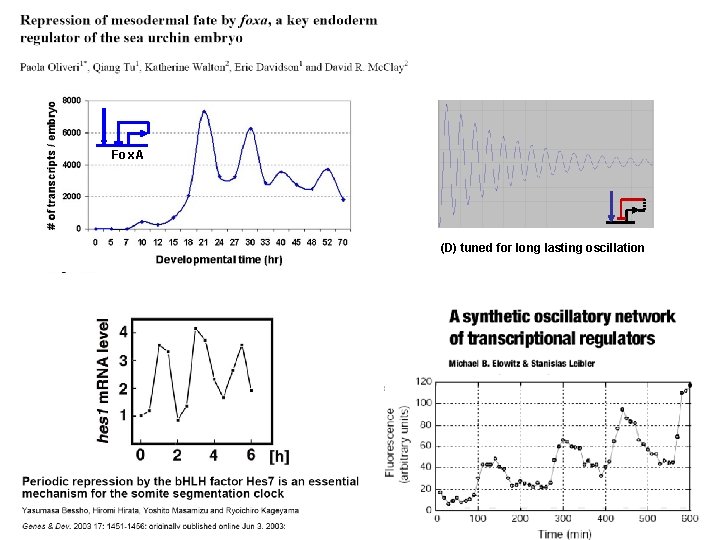
Fox. A (D) tuned for long lasting oscillation

Potential genetic regulatory functional building blocks (D) (A) (E) (F) (B) (C) (g)


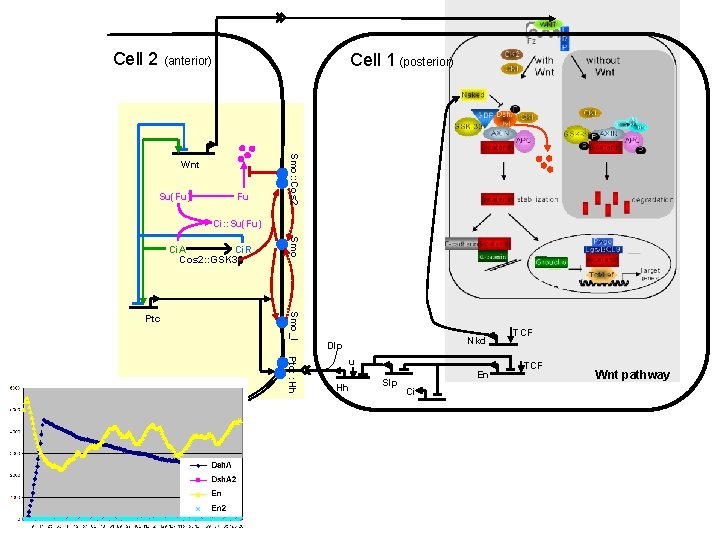
Cell 2 (anterior) Cell 1 (posterior) Su(Fu) Fu Smo: : Cos 2 Wnt Ci: : Su(Fu) Ptc: : Hh Hh pathway Smo_I Ptc Smo Ci. A Ci. R Cos 2: : GSK 3 b Nkd Dlp u Hh Slp En Ci TCF Wnt pathway

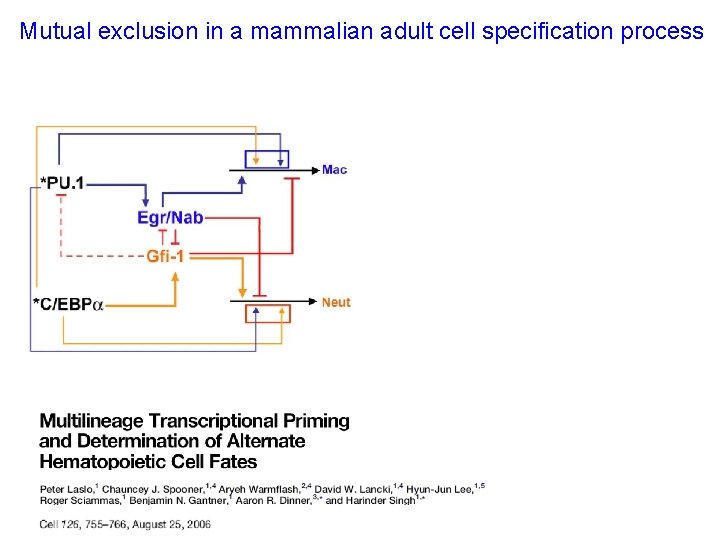
Mutual exclusion in a mammalian adult cell specification process

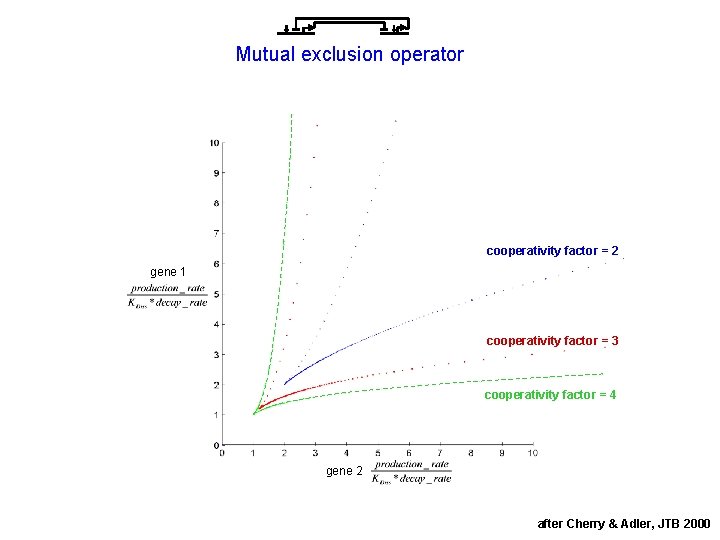
Mutual exclusion operator cooperativity factor = 2 gene 1 cooperativity factor = 3 cooperativity factor = 4 gene 2 after Cherry & Adler, JTB 2000

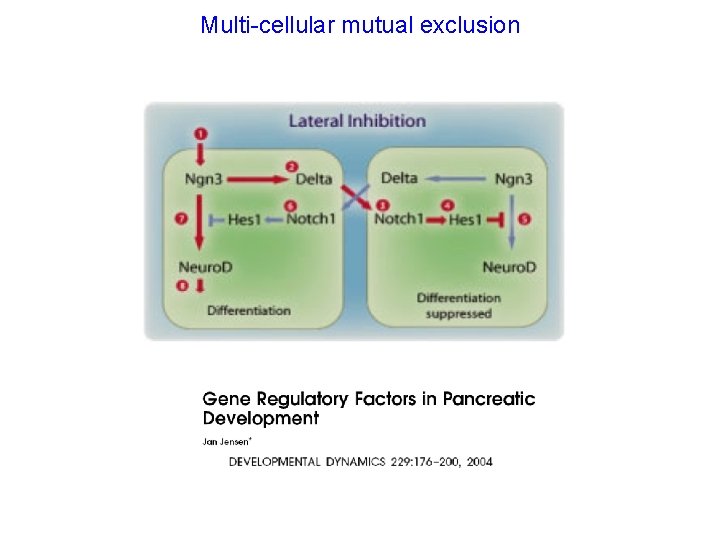
Multi-cellular mutual exclusion

Potential genetic regulatory functional building blocks (D) (A) (E) (F) (B) (C) (g)

The feed forward motif: Alon & colleagues, Nature Genetics, 2002. 31(1): p. 64 -8 see also:

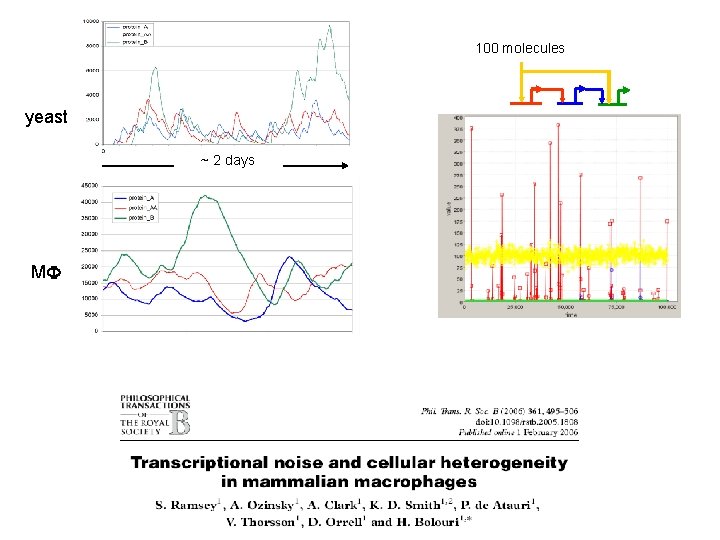
100 molecules yeast ~ 2 days MF

Potential genetic regulatory functional building blocks (D) (A) (E) (F) (B) (C) (g)


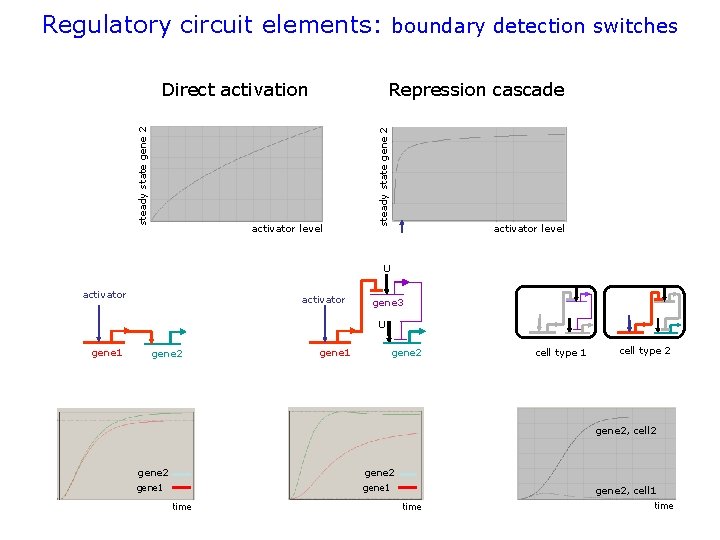
Regulatory circuit elements: boundary detection switches Repression cascade activator level steady state gene 2 Direct activation activator level U activator gene 3 U gene 1 gene 2 cell type 1 cell type 2 gene 2, cell 2 gene 1 gene 2 gene 1 time gene 2, cell 1 time


Potential genetic regulatory functional building blocks (D) (A) (E) (F) (B) (C) (g)



Potential genetic regulatory functional building blocks (D) (A) (E) (F) (B) (C) (g)

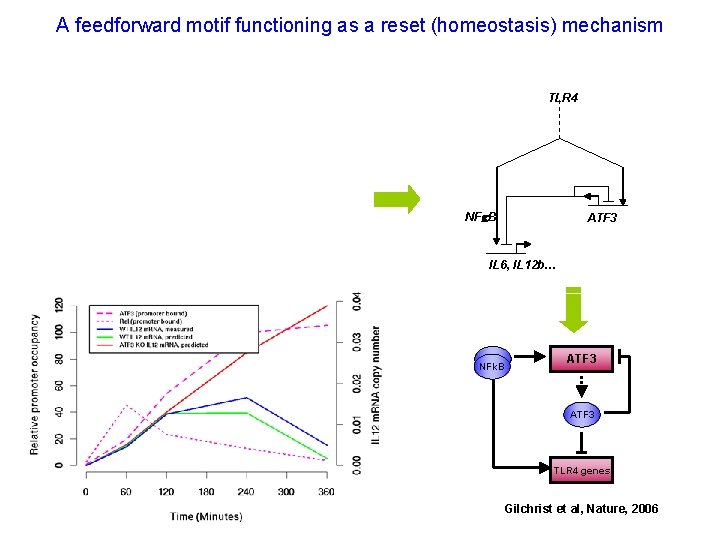
A feedforward motif functioning as a reset (homeostasis) mechanism TLR 4 NFk. B ATF 3 IL 6, IL 12 b… NFk. B ATF 3 TLR 4 genes Gilchrist et al, Nature, 2006


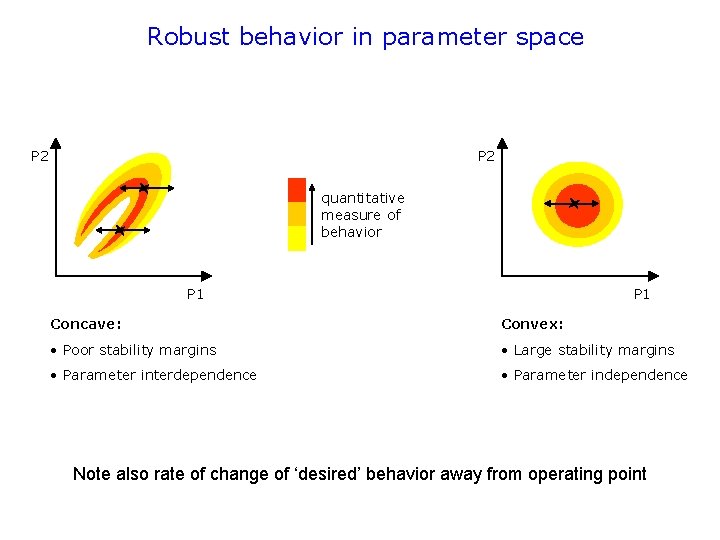
Robust behavior in parameter space P 2 quantitative measure of behavior P 1 Concave: Convex: • Poor stability margins • Large stability margins • Parameter interdependence • Parameter independence Note also rate of change of ‘desired’ behavior away from operating point

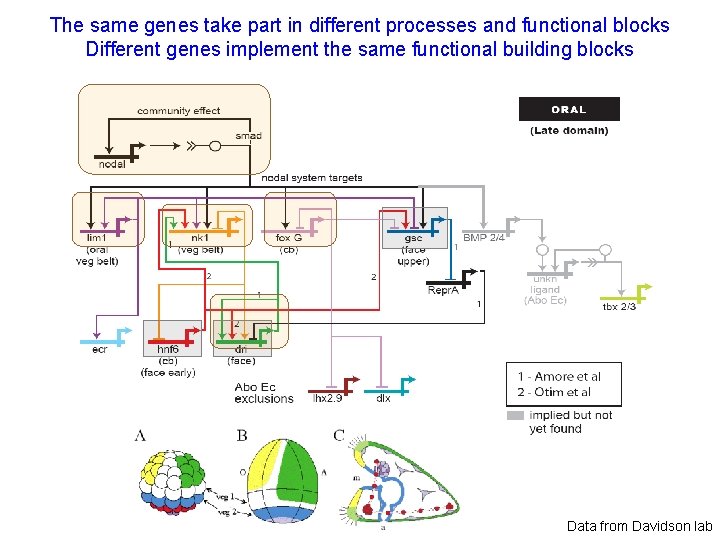
The same genes take part in different processes and functional blocks Different genes implement the same functional building blocks Data from Davidson lab

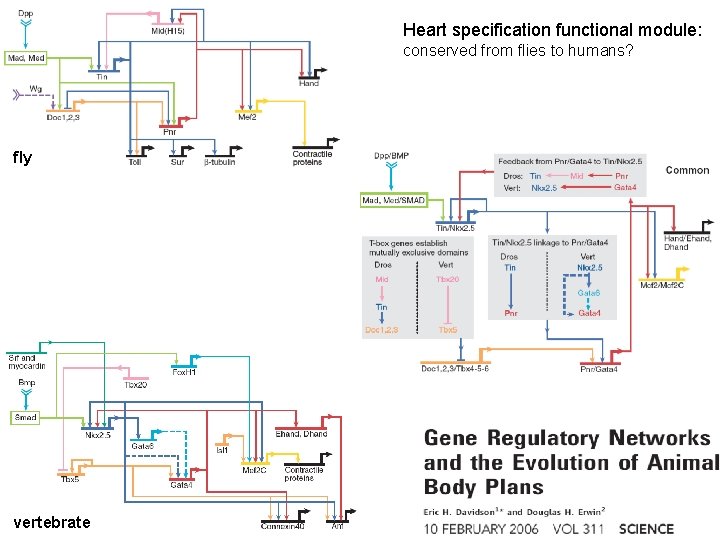
Heart specification functional module: conserved from flies to humans? fly vertebrate


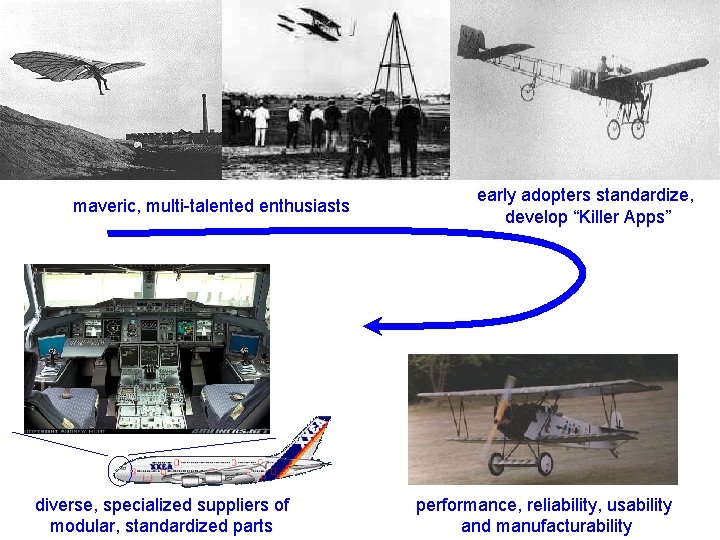
maveric, multi-talented enthusiasts diverse, specialized suppliers of modular, standardized parts early adopters standardize, develop “Killer Apps” performance, reliability, usability and manufacturability

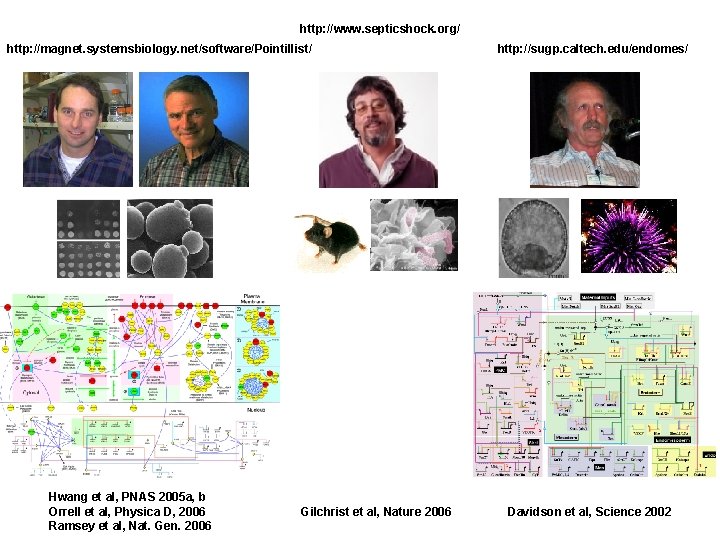
http: //www. septicshock. org/ http: //magnet. systemsbiology. net/software/Pointillist/ Hwang et al, PNAS 2005 a, b Orrell et al, Physica D, 2006 Ramsey et al, Nat. Gen. 2006 Gilchrist et al, Nature 2006 http: //sugp. caltech. edu/endomes/ Davidson et al, Science 2002

Davidson Lab & Eric Davidson Bill Longabaugh Jennifer Smith, Deena Leslie, Andrea Weston, Marcello Marelli, Tim Petersen Andy Siegel, John Aitchison, Lee Hood Ben Buelow, Mark Gilchrist, Katy Kennedy, Adrian Ozinsky, Jared Roach, Carrie Baldwin, Natalya Yudkovsly Alan Aderem Steve Ramsey David Orrell Alistair Rust Martin Korb Vesteinn Thorsson Daehee Hwang Pedro de Atauri Mark Robinson Christophe Battail Bin Li

Department of Cellular & Physiological Sciences University of British Columbia Life Sciences Centre 2350 Health Sciences Mall Vancouver, BC Canada V 6 T 1 Z 3 Recruiting: - Graduate students - Post docs - Technicians - Professional staff Subjects: - molecular & cell biology - transcriptional regulation - single cell assays - math, stats - physics, engineering - computer science - software engineering