This work is licensed under a Creative Commons




- Slides: 39

This work is licensed under a Creative Commons Attribution-Non. Commercial-Share. Alike License. Your use of this material constitutes acceptance of that license and the conditions of use of materials on this site. Copyright 2006, The Johns Hopkins University and Sean T. Prigge. All rights reserved. Use of these materials permitted only in accordance with license rights granted. Materials provided “AS IS”; no representations or warranties provided. User assumes all responsibility for use, and all liability related thereto, and must independently review all materials for accuracy and efficacy. May contain materials owned by others. User is responsible for obtaining permissions for use from third parties as needed.

New Drug Targets Sean T. Prigge, Ph. D

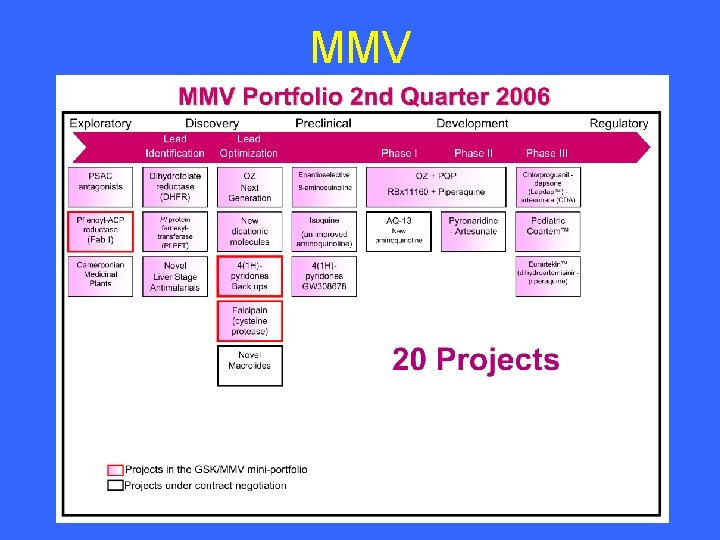
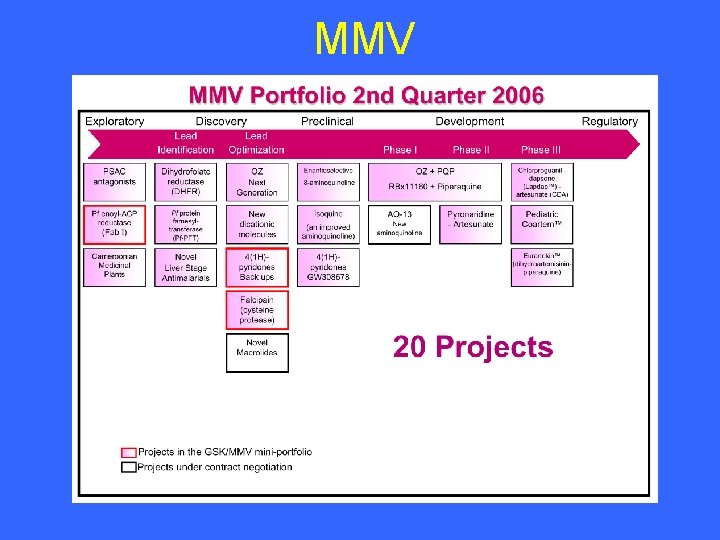
MMV

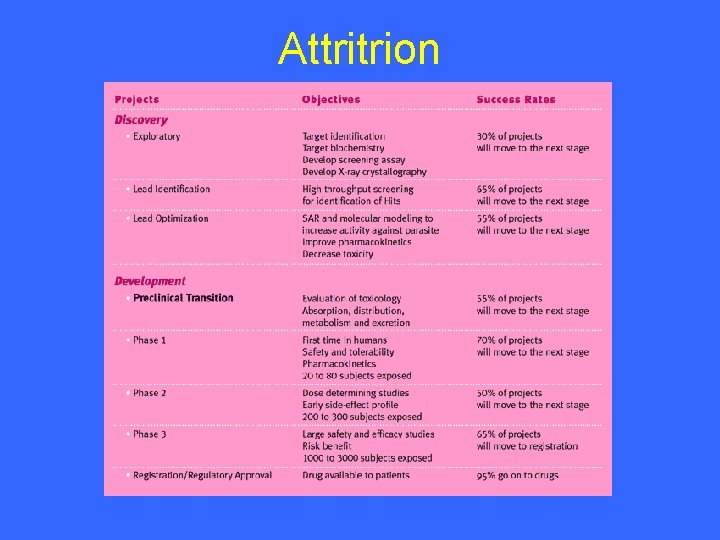
Attritrion

Attrition There are several reasons for the high dropout rate: u a biologically poor target u lack of activity against the target or parasite u toxicity u tolerability u cost of goods

MMV

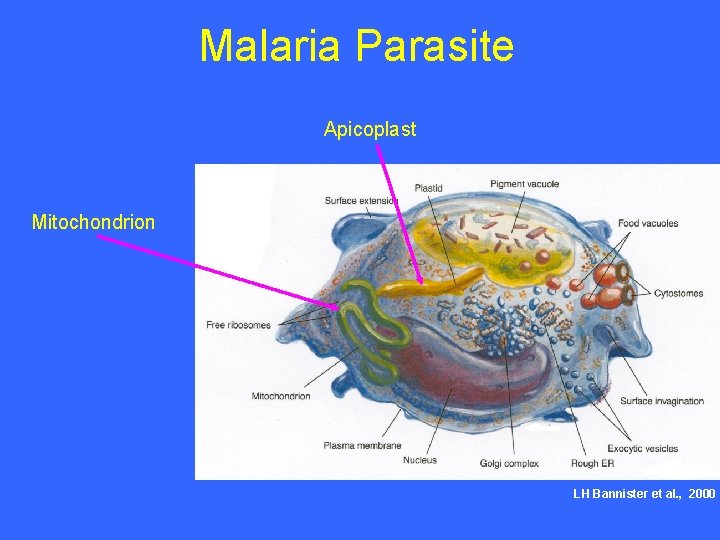
Malaria Parasite Apicoplast Mitochondrion LH Bannister et al. , 2000

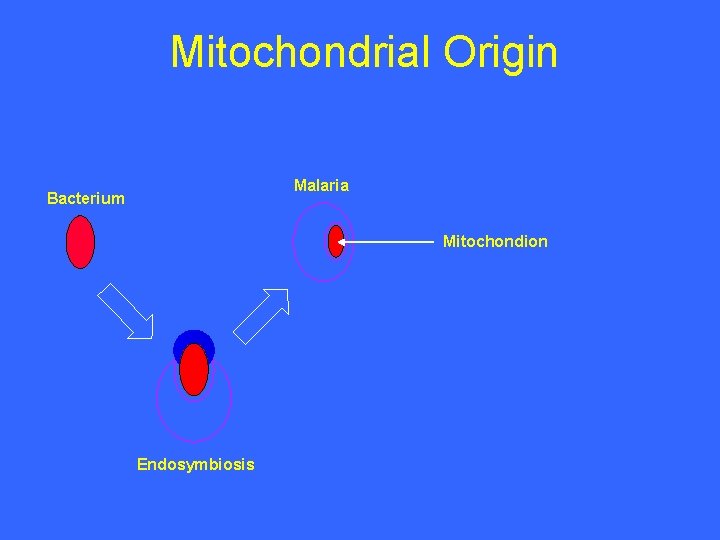
Mitochondrial Origin Malaria Bacterium Mitochondion Endosymbiosis

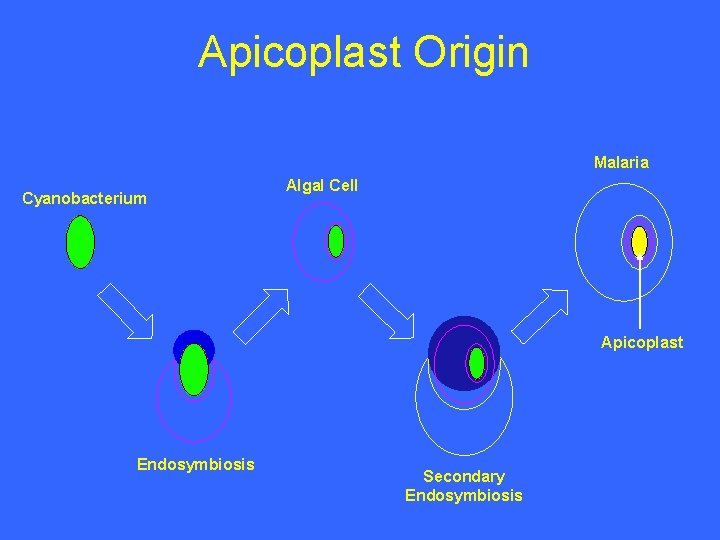
Apicoplast Origin Malaria Cyanobacterium Algal Cell Apicoplast Endosymbiosis Secondary Endosymbiosis

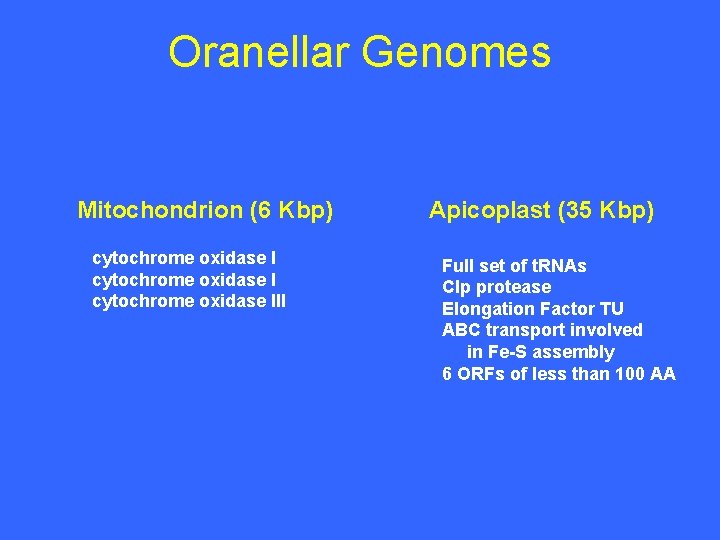
Oranellar Genomes Mitochondrion (6 Kbp) cytochrome oxidase III Apicoplast (35 Kbp) Full set of t. RNAs Clp protease Elongation Factor TU ABC transport involved in Fe-S assembly 6 ORFs of less than 100 AA

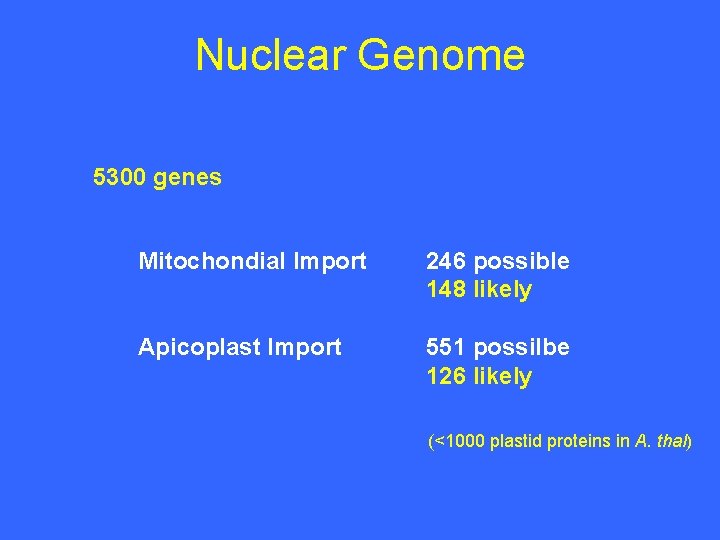
Nuclear Genome 5300 genes Mitochondial Import 246 possible 148 likely Apicoplast Import 551 possilbe 126 likely (<1000 plastid proteins in A. thal)

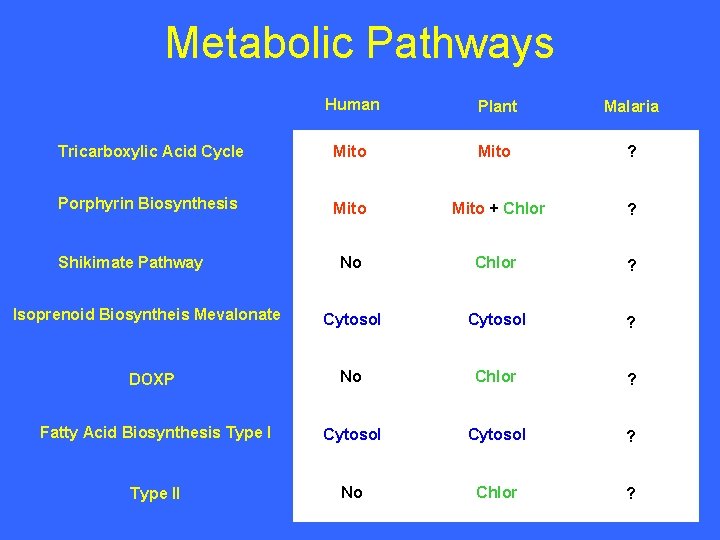
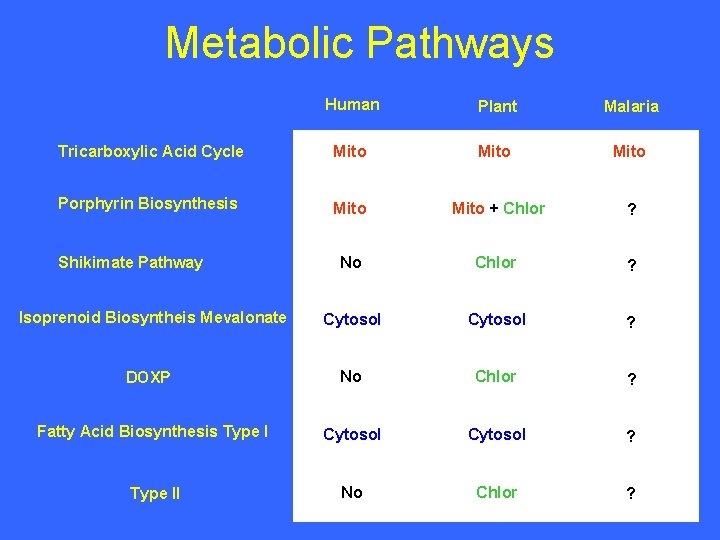
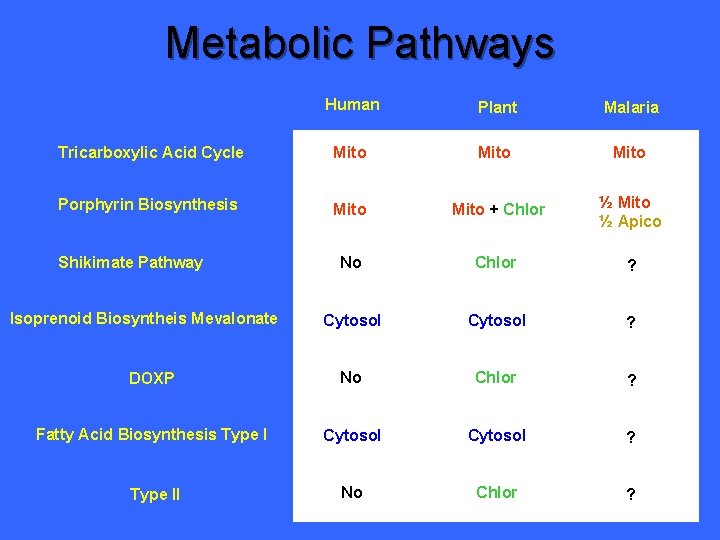
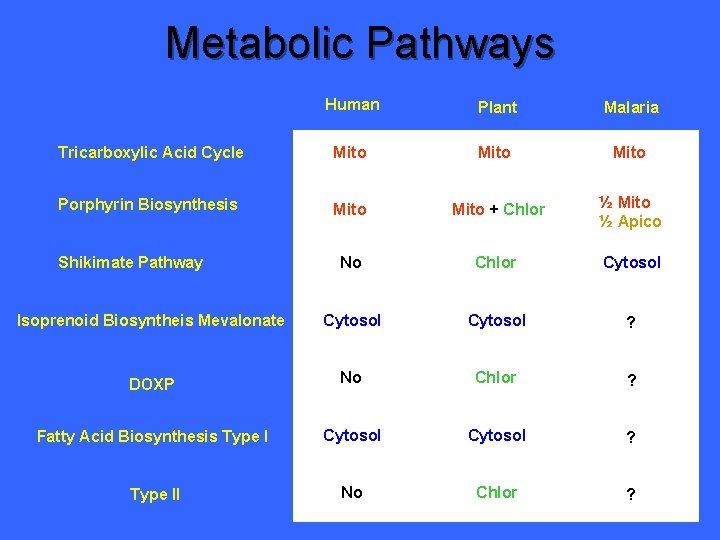
Metabolic Pathways Human Plant Malaria Tricarboxylic Acid Cycle Mito ? Porphyrin Biosynthesis Mito + Chlor ? No Chlor ? Cytosol ? DOXP No Chlor ? Fatty Acid Biosynthesis Type I Cytosol ? Type II No Chlor ? Shikimate Pathway Isoprenoid Biosyntheis Mevalonate

http: //www. genome. jp/kegg/pathway. html

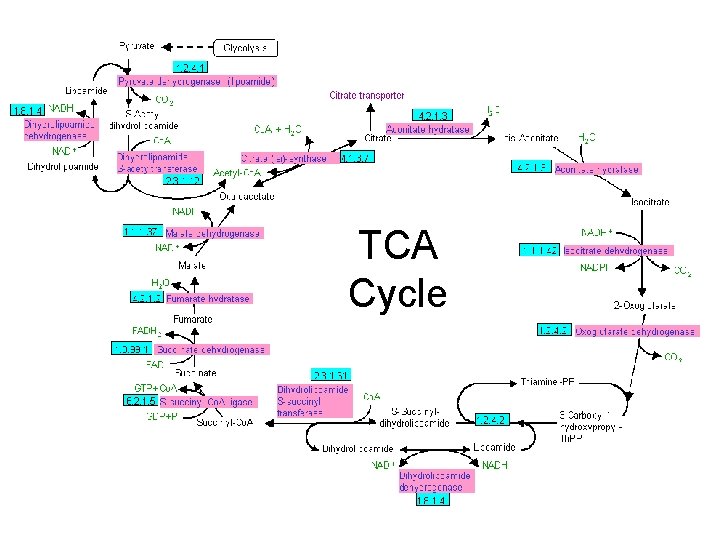
TCA Cycle

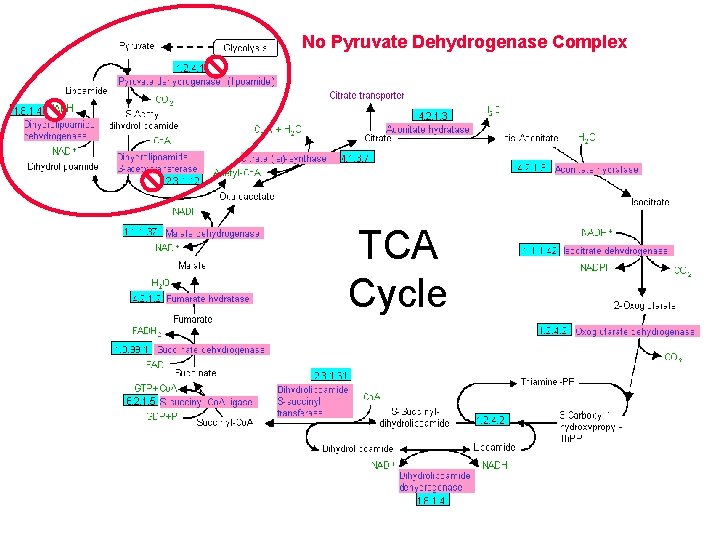
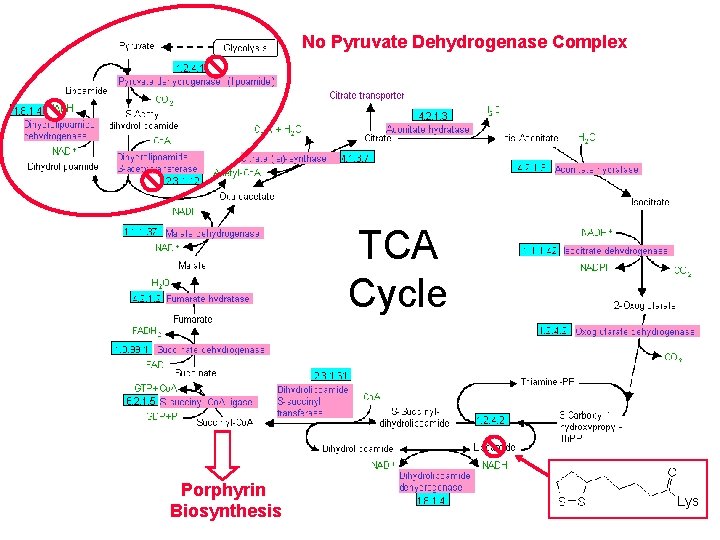
No Pyruvate Dehydrogenase Complex TCA Cycle

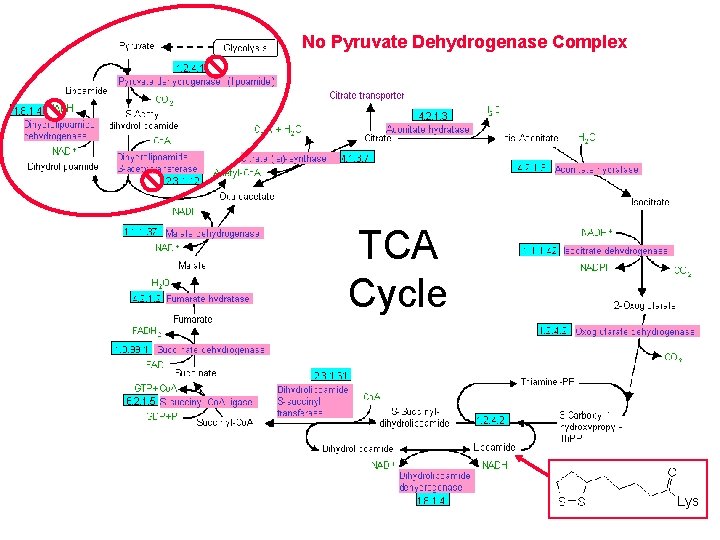
No Pyruvate Dehydrogenase Complex TCA Cycle Lys

No Pyruvate Dehydrogenase Complex TCA Cycle Porphyrin Biosynthesis Lys

Metabolic Pathways Human Plant Malaria Tricarboxylic Acid Cycle Mito Porphyrin Biosynthesis Mito + Chlor ? No Chlor ? Isoprenoid Biosyntheis Mevalonate Cytosol ? DOXP No Chlor ? Fatty Acid Biosynthesis Type I Cytosol ? Type II No Chlor ? Shikimate Pathway

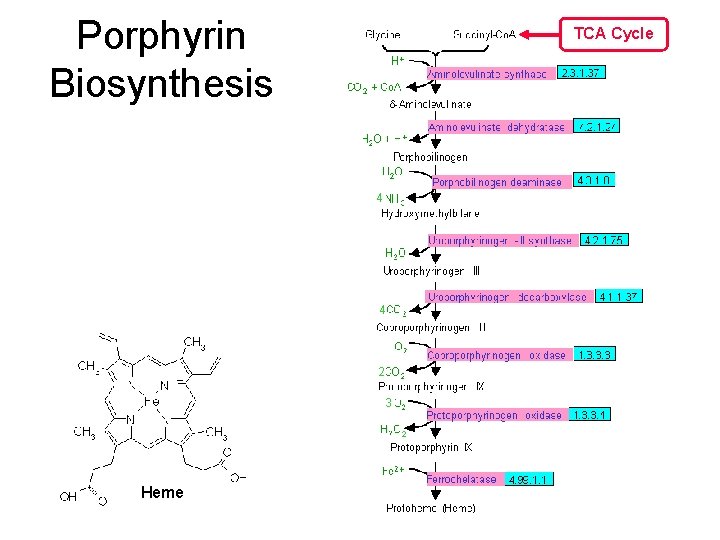
Porphyrin Biosynthesis TCA Cycle 4 4 2 H 2 O + 2 3 3 Heme 2

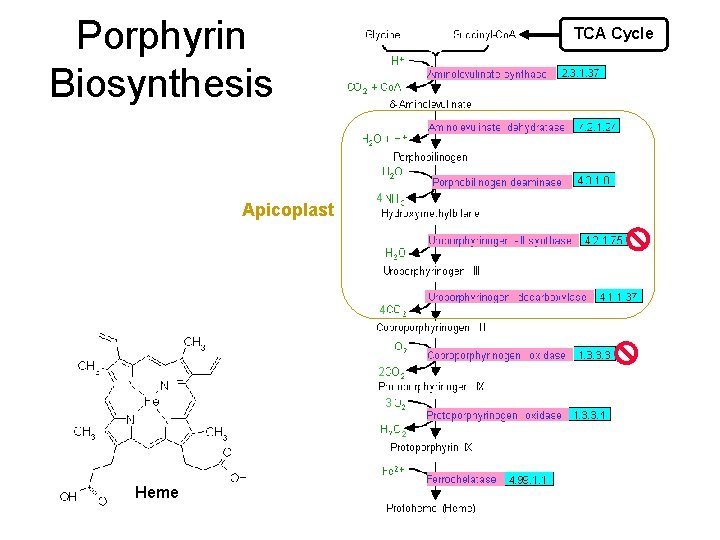
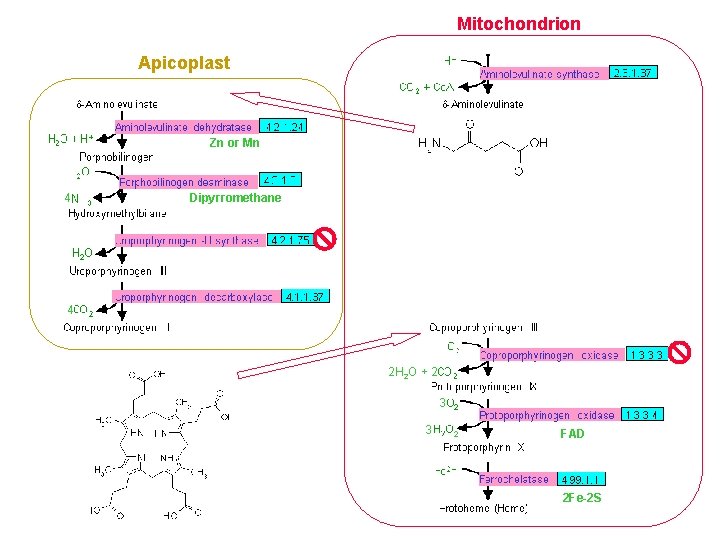
Porphyrin Biosynthesis Apicoplast TCA Cycle 4 4 2 H 2 O + 2 3 3 Heme 2

Mitochondrion TCA Cycle Apicoplast Zn or Mn 4 Dipyrromethane 4 2 H 2 O + 2 3 3 2 FAD 2 Fe-2 S TCA Cycle

Metabolic Pathways Human Plant Malaria Tricarboxylic Acid Cycle Mito Porphyrin Biosynthesis Mito + Chlor ½ Mito ½ Apico No Chlor ? Cytosol ? DOXP No Chlor ? Fatty Acid Biosynthesis Type I Cytosol ? Type II No Chlor ? Shikimate Pathway Isoprenoid Biosyntheis Mevalonate

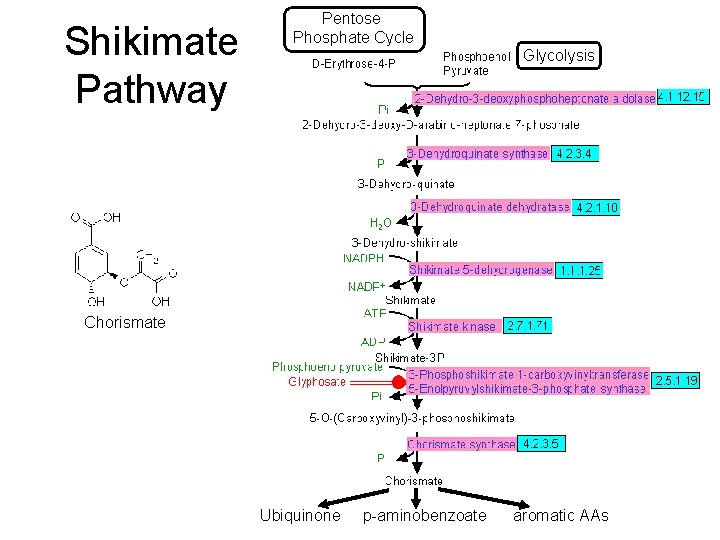
Shikimate Pathway Pentose Phosphate Cycle Glycolysis Chorismate Ubiquinone p-aminobenzoate aromatic AAs

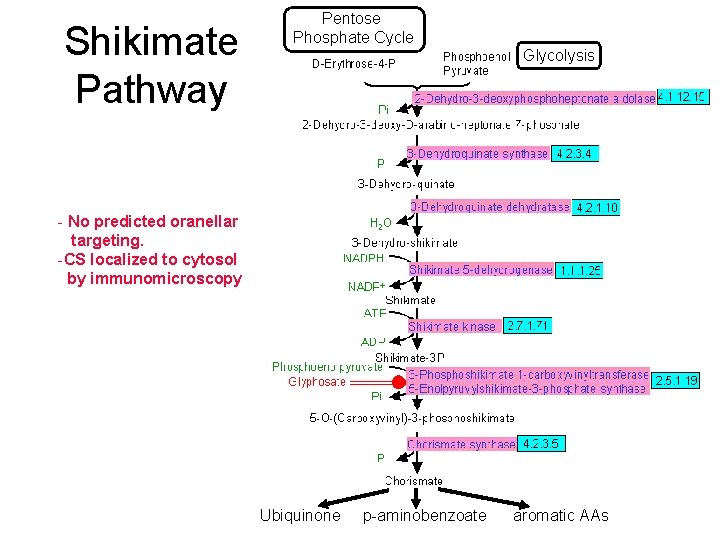
Shikimate Pathway Pentose Phosphate Cycle Glycolysis - No predicted oranellar targeting. -CS localized to cytosol by immunomicroscopy Ubiquinone p-aminobenzoate aromatic AAs

Metabolic Pathways Human Plant Malaria Tricarboxylic Acid Cycle Mito Porphyrin Biosynthesis Mito + Chlor ½ Mito ½ Apico No Chlor Cytosol Isoprenoid Biosyntheis Mevalonate Cytosol ? DOXP No Chlor ? Fatty Acid Biosynthesis Type I Cytosol ? Type II No Chlor ? Shikimate Pathway

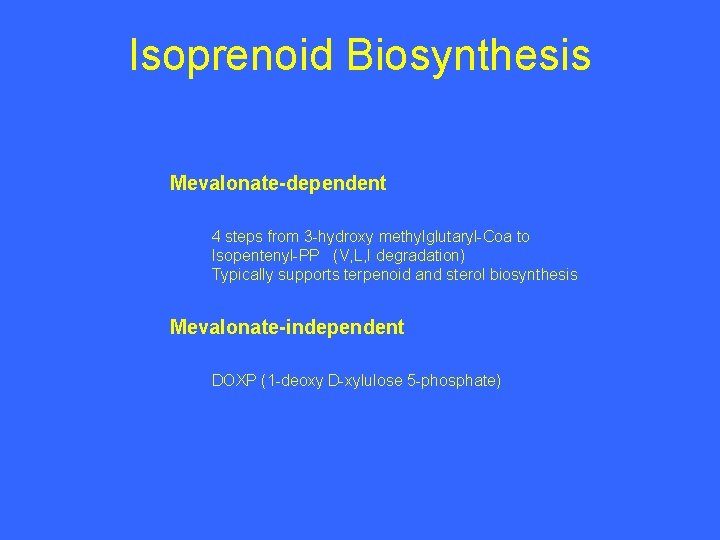
Isoprenoid Biosynthesis Mevalonate-dependent 4 steps from 3 -hydroxy methylglutaryl-Coa to Isopentenyl-PP (V, L, I degradation) Typically supports terpenoid and sterol biosynthesis Mevalonate-independent DOXP (1 -deoxy D-xylulose 5 -phosphate)

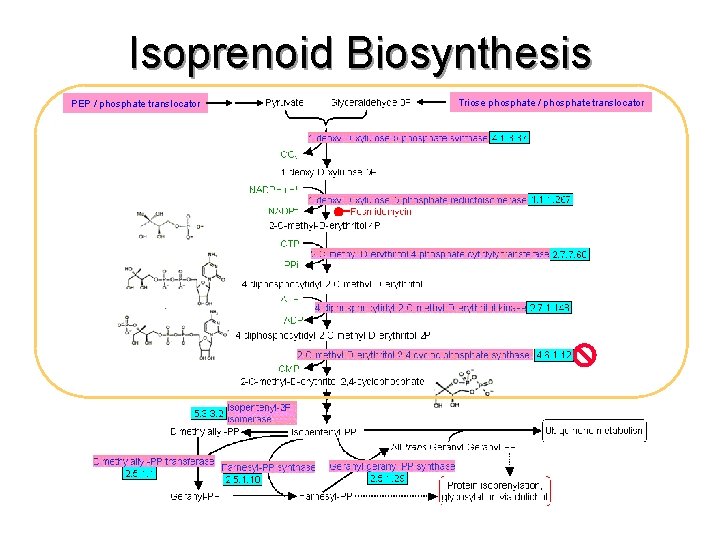
Isoprenoid Biosynthesis PEP / phosphate translocator Triose phosphate / phosphate translocator

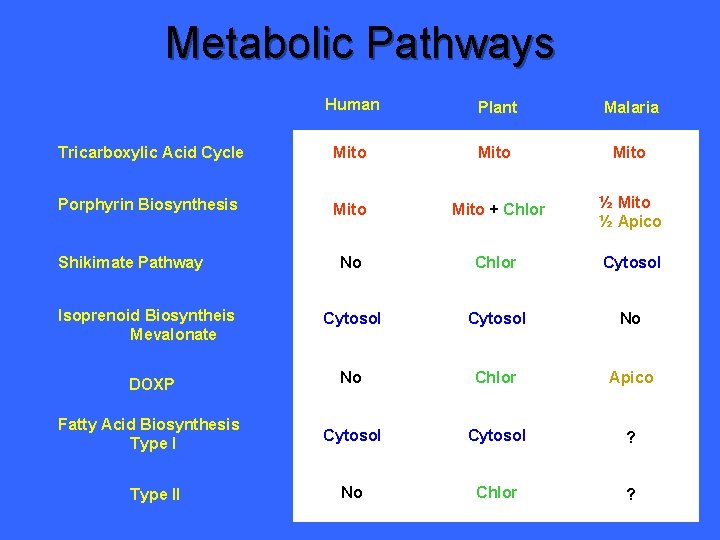
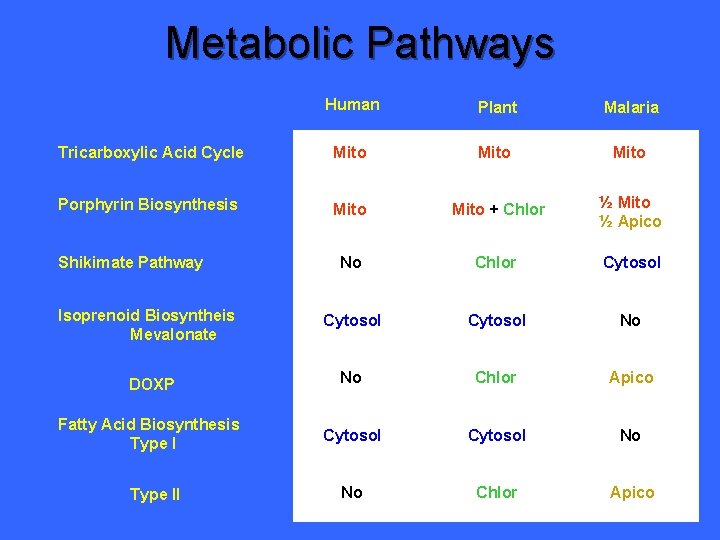
Metabolic Pathways Human Plant Malaria Tricarboxylic Acid Cycle Mito Porphyrin Biosynthesis Mito + Chlor ½ Mito ½ Apico No Chlor Cytosol No DOXP No Chlor Apico Fatty Acid Biosynthesis Type I Cytosol ? No Chlor ? Shikimate Pathway Isoprenoid Biosyntheis Mevalonate Type II

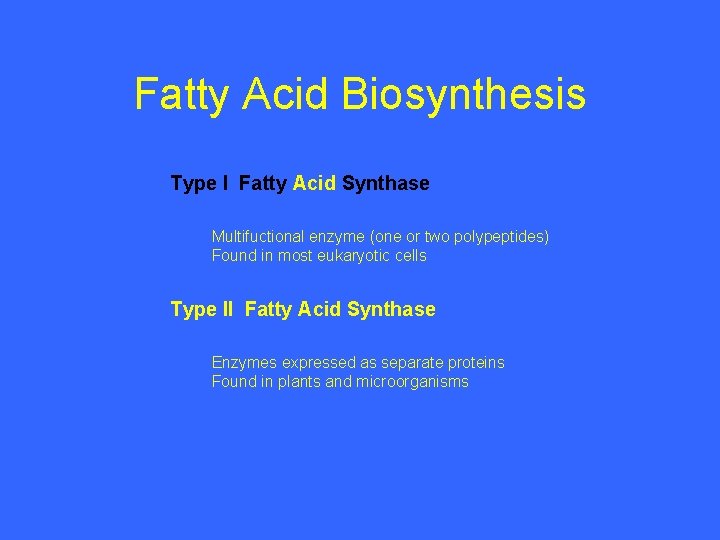
Fatty Acid Biosynthesis Type I Fatty Acid Synthase Multifuctional enzyme (one or two polypeptides) Found in most eukaryotic cells Type II Fatty Acid Synthase Enzymes expressed as separate proteins Found in plants and microorganisms

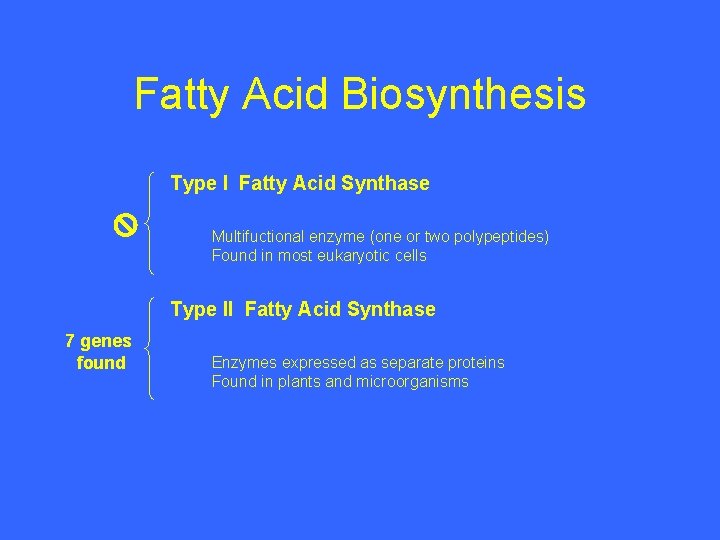
Fatty Acid Biosynthesis Type I Fatty Acid Synthase Multifuctional enzyme (one or two polypeptides) Found in most eukaryotic cells Type II Fatty Acid Synthase 7 genes found Enzymes expressed as separate proteins Found in plants and microorganisms

Type II FAS Kyoto Encyclopedia of Genes and Genomes

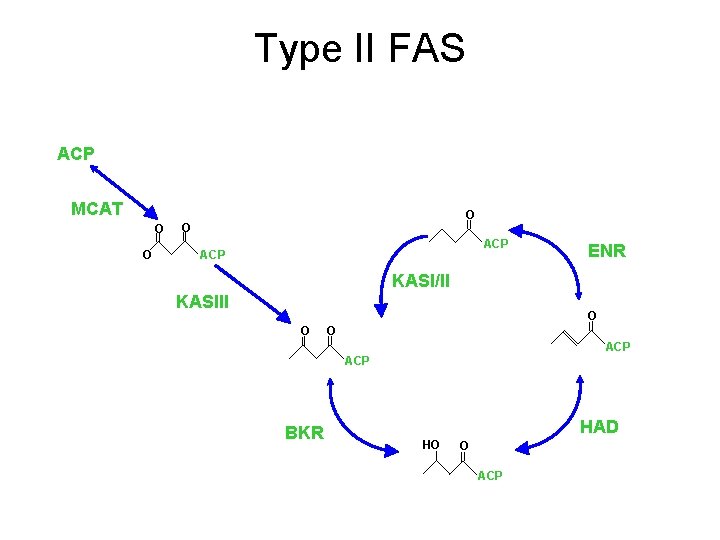
Type II FAS ACP MCAT O O ? ACP ENR KASI/II KASIII O O O ACP BKR HAD HO O ACP

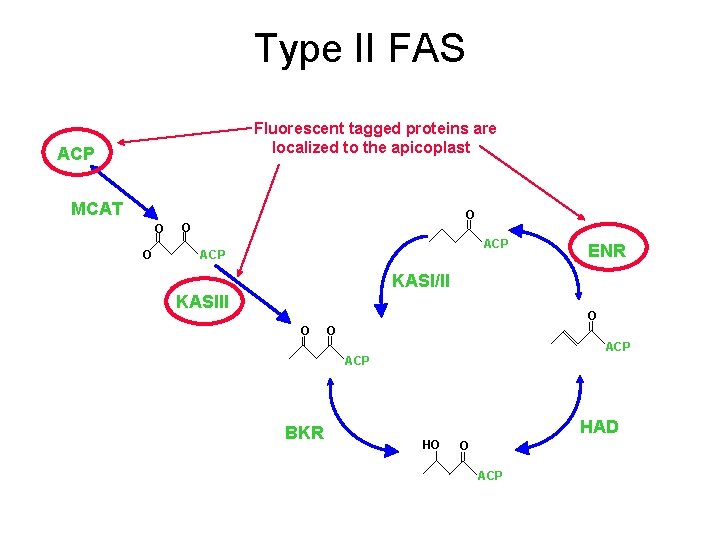
Type II FAS Fluorescent tagged proteins are localized to the apicoplast ACP MCAT O O ? ACP ENR KASI/II KASIII O O O ACP BKR HAD HO O ACP

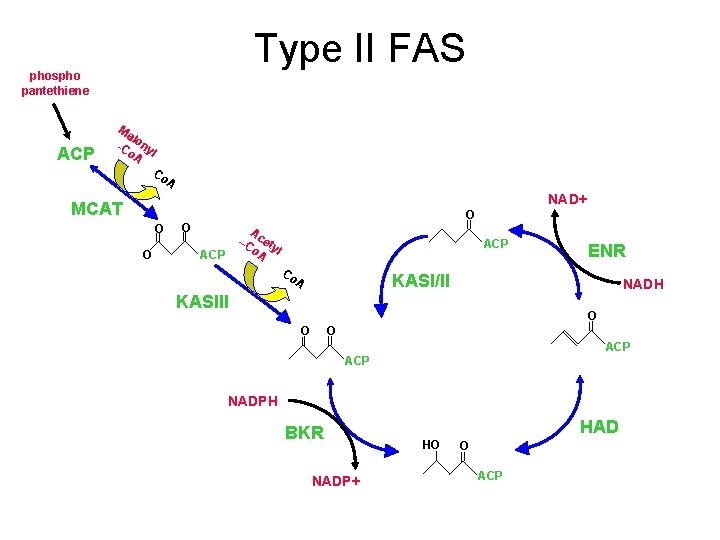
Type II FAS phospho pantethiene ACP Ma l -C ony o. A l Co A NAD+ MCAT O O ? ACP Co A ENR KASI/II NADH KASIII O O O ACP NADPH BKR NADP+ HAD HO O A --C cety o. A l ACP

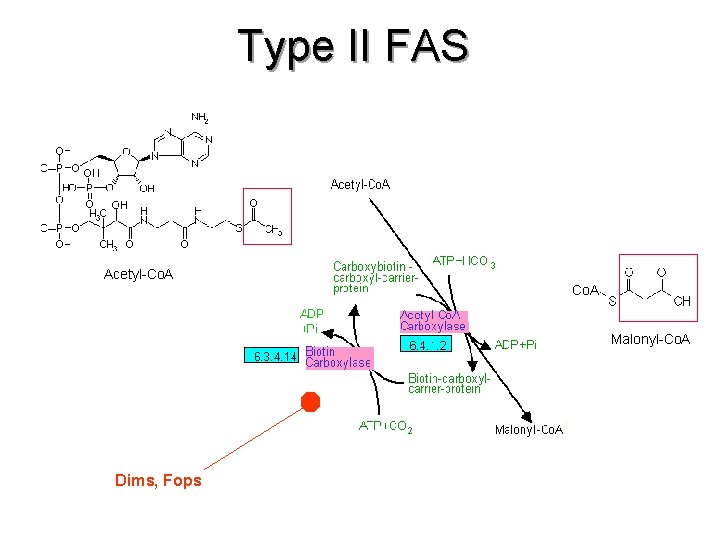
Type II FAS Acetyl-Co. A Malonyl-Co. A Dims, Fops

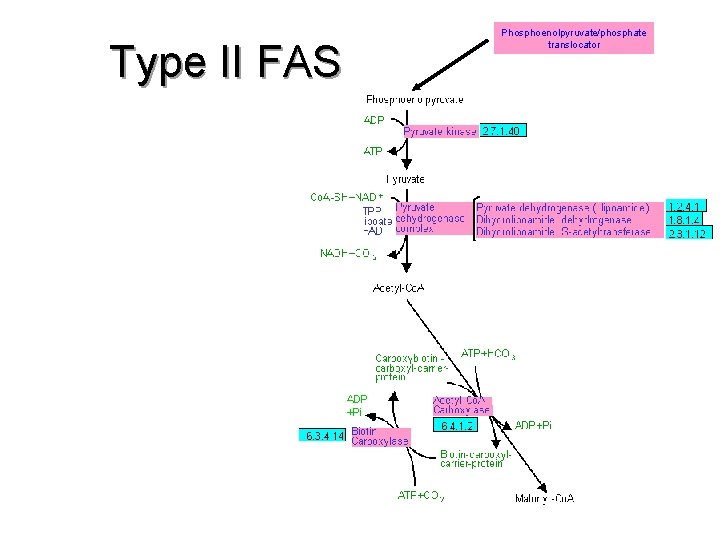
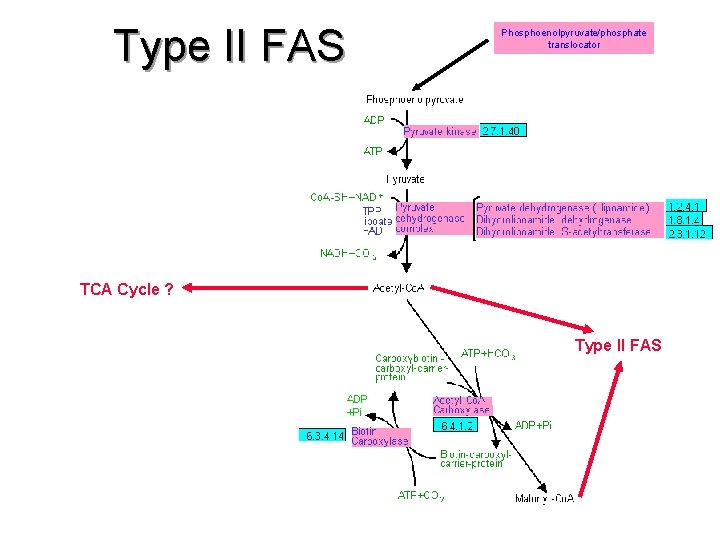
Type II FAS Phosphoenolpyruvate/phosphate translocator

Type II FAS Phosphoenolpyruvate/phosphate translocator TCA Cycle ? Type II FAS

Metabolic Pathways Human Plant Malaria Tricarboxylic Acid Cycle Mito Porphyrin Biosynthesis Mito + Chlor ½ Mito ½ Apico No Chlor Cytosol No DOXP No Chlor Apico Fatty Acid Biosynthesis Type I Cytosol No No Chlor Apico Shikimate Pathway Isoprenoid Biosyntheis Mevalonate Type II

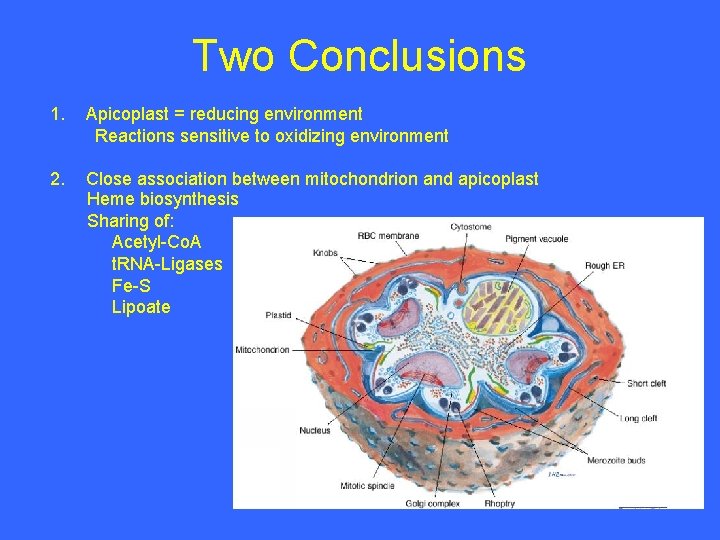
Two Conclusions 1. Apicoplast = reducing environment Reactions sensitive to oxidizing environment 2. Close association between mitochondrion and apicoplast Heme biosynthesis Sharing of: Acetyl-Co. A t. RNA-Ligases Fe-S Lipoate