The Thicket of Challenges in GPCR Molecular Pharmacology











![[ligand] T T T %Bound unfused [ligand] T T Transducer • This would require [ligand] T T T %Bound unfused [ligand] T T Transducer • This would require](https://slidetodoc.com/presentation_image/606e74e499ae9db3b1e7947f54d0e8fb/image-27.jpg)



- Slides: 31

The Thicket of Challenges in GPCR Molecular Pharmacology: Paradigm shifts bring new opportunities and new hurdles Ryan T. Strachan, Ph. D

Goals: • Convey my enthusiasm about the current and future states of GPCR Drug Discovery • Includes the first presentation of a novel screening strategy aimed at studying the unexplored pharmacology of GPCRs (screening the ‘transducerome’) • Initiate a discussion about how to best apply computational methods to facilitate GPCR drug discovery (if at all) • Establish strategic collaborations to facilitate the generation of robust and predictive Deep Learning methods

The role of the Molecular Pharmacologist in GPCR Drug Discovery: • Concerned with the study of drug action on a molecular and chemical level • Seek to discover and validate new therapeutic strategies to improve human health • Draw from ideas across multiple fields of study: • • Biochemistry Chemistry Physiology Computer Science • • Clinical Medicine Mathematics Engineering Statistics

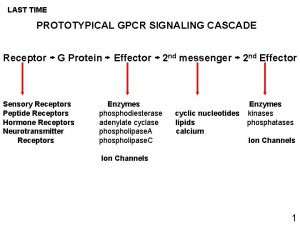
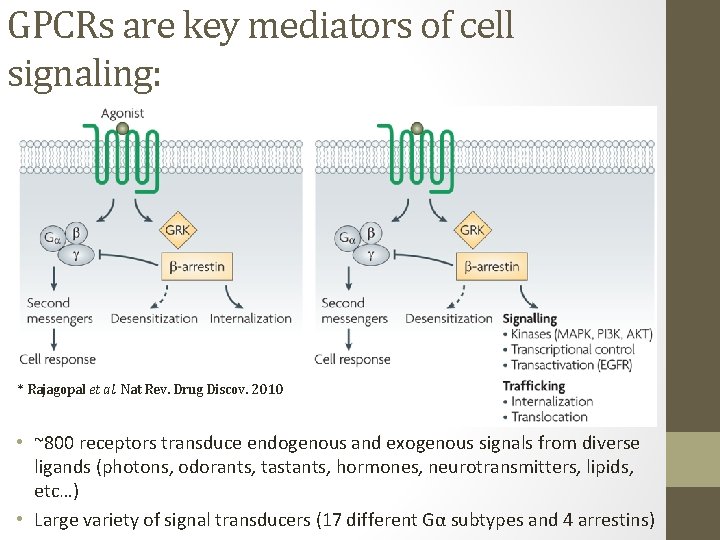
GPCRs are key mediators of cell signaling: * Rajagopal et al. Nat Rev. Drug Discov. 2010 • ~800 receptors transduce endogenous and exogenous signals from diverse ligands (photons, odorants, tastants, hormones, neurotransmitters, lipids, etc…) • Large variety of signal transducers (17 different Gα subtypes and 4 arrestins)

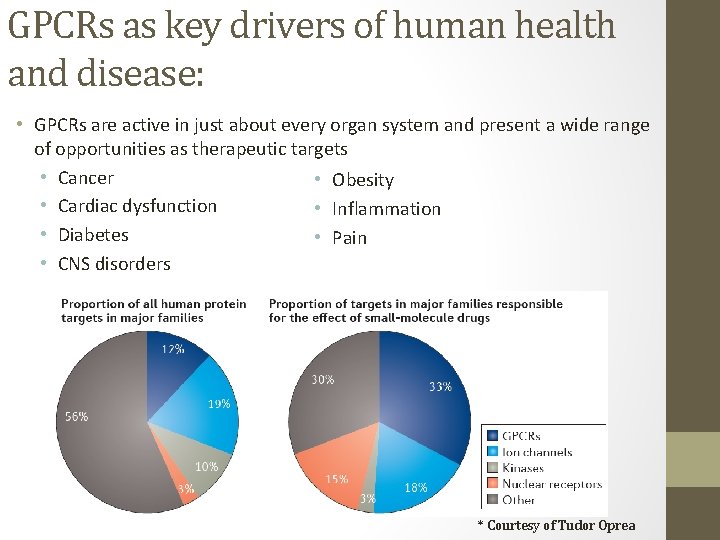
GPCRs as key drivers of human health and disease: • GPCRs are active in just about every organ system and present a wide range of opportunities as therapeutic targets • Cancer • Obesity • Cardiac dysfunction • Inflammation • Diabetes • Pain • CNS disorders * Courtesy of Tudor Oprea

Classic theories give way to new paradigms: • As key drivers of human (patho)physiology GPCRs have been widely studied 1900’s - ‘Chemoreceptors’ and mathematical models of signaling added order to chaos 19701990’s - Advent of functional assays, radioligand binding assays, and the cloning of G proteins and GPCRs paved the way for biophysical and structure-function studies 2000 - Sequencing of the human genome revealed the full complement of GPCRs, including the identification of orphan GPCRs 1990’spresent - Advances in assay technology revealed that GPCR agonists disproportionately activate numerous cellular pathways (ie. , biased agonism) via unique receptor conformations 2007 present - Advances in GPCR crystallography and computational approaches are ushering in a Golden Age of Molecular Pharmacology, launching the field of structure-based drug discovery

Not so fast, we have a long way to go….

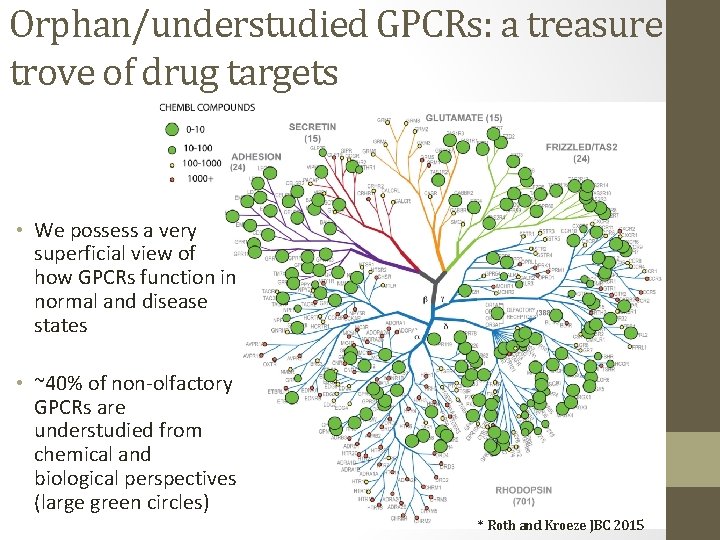
Orphan/understudied GPCRs: a treasure trove of drug targets • We possess a very superficial view of how GPCRs function in normal and disease states • ~40% of non-olfactory GPCRs are understudied from chemical and biological perspectives (large green circles) * Roth and Kroeze JBC 2015

Opportunities presented by orphan GPCRs: • Well-characterized GPCRs play key roles in (patho)physiology, therefore orphan/understudied GPCRs have untapped therapeutic potential • Small molecule receptors: • G-21 (5 -HT 1 A serotonin) • RGB-2 (D 2 dopamine) • Neuropeptide receptors: • ORL-1 (Orphanin. FQ/Nociceptin) • HFGAN 72 (Orexin 1) • GPR 10 (Prolactin-releasing peptide) • APJ (Apelin) • GHSR (Ghrelin) • GPR 54 (Kisspeptin/metastin) • GPR 73 a/b (Prokineticin) • GPR 154 (Neuropeptide S) * Civelli et al. Ann. Rev. Pharmacol. Toxicol. 2013

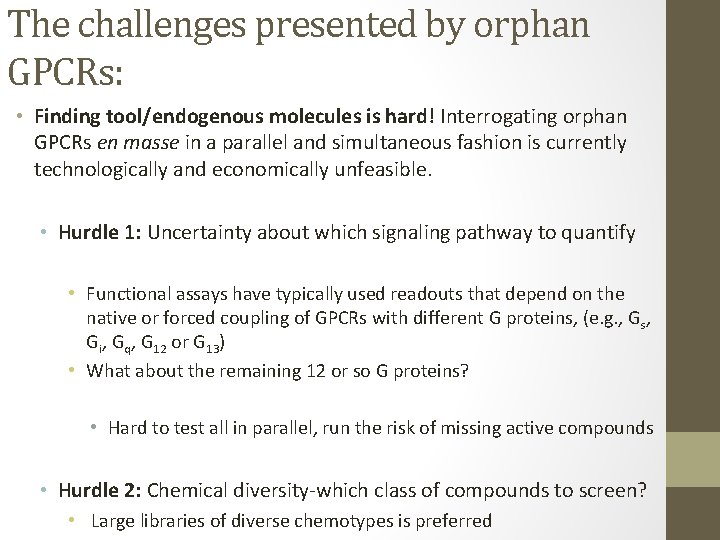
The challenges presented by orphan GPCRs: • Finding tool/endogenous molecules is hard! Interrogating orphan GPCRs en masse in a parallel and simultaneous fashion is currently technologically and economically unfeasible. • Hurdle 1: Uncertainty about which signaling pathway to quantify • Functional assays have typically used readouts that depend on the native or forced coupling of GPCRs with different G proteins, (e. g. , Gs, Gi, Gq, G 12 or G 13) • What about the remaining 12 or so G proteins? • Hard to test all in parallel, run the risk of missing active compounds • Hurdle 2: Chemical diversity-which class of compounds to screen? • Large libraries of diverse chemotypes is preferred

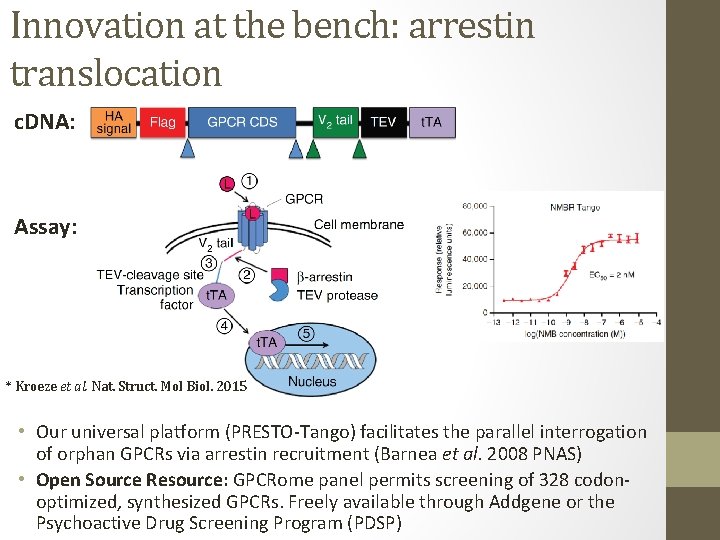
Innovation at the bench: arrestin translocation c. DNA: Assay: * Kroeze et al. Nat. Struct. Mol Biol. 2015 • Our universal platform (PRESTO-Tango) facilitates the parallel interrogation of orphan GPCRs via arrestin recruitment (Barnea et al. 2008 PNAS) • Open Source Resource: GPCRome panel permits screening of 328 codonoptimized, synthesized GPCRs. Freely available through Addgene or the Psychoactive Drug Screening Program (PDSP)

Integrating physical/computational methods to overcome the hurdles of orphan GPCR screening: • Addressed the issue of chemical diversity by integrating physical/computational methods to facilitate tool molecule identification from tens of millions of virtual compounds • Part of a larger discovery effort by the NIH to ‘Illuminate the Druggable Genome (IDG) (https: //commonfund. nih. gov/idg/index) • Develop novel, scalable technologies to shed light on the ‘dark matter’ of the human genome in an effort to identify new biology and new therapies • Ion channels • Nuclear receptors • Kinases * Opportunities to mine these datasets via Deep Learning?

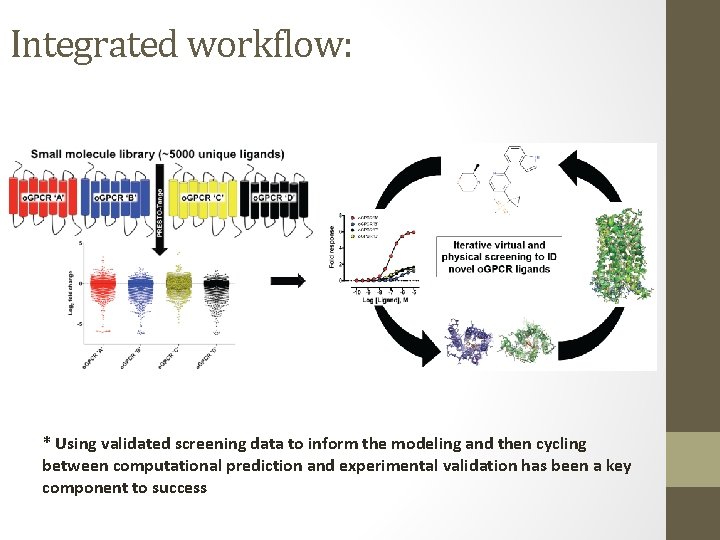
Integrated workflow: * Using validated screening data to inform the modeling and then cycling between computational prediction and experimental validation has been a key component to success

Integrating physical and computational approaches identifies novel chemical matter to reveal new biology: • GPR 68 (Huang et al. Nature. 2016) • Proton-sensing GPCR, understudied and lacks tool molecules • Identified a small molecule positive allosteric modulator (PAM), Ogerin • GPR 65 (Huang et al. Nature. 2016) • Proton-sensing GPCR with 37% identity to GPR 68, understudied • Identified an allosteric agonist and a negative allosteric modulator (NAM) • MRGPRX 2 (Lansu et al. Nat. Chem. Biol. in review) • Understudied primate-exclusive GPCR associated with pain and itch • Identified a selective submicromolar agonist tool compound

A second major paradigm shift is ‘biased agonism’, which is revolutionizing how we target GPCRs with drugs

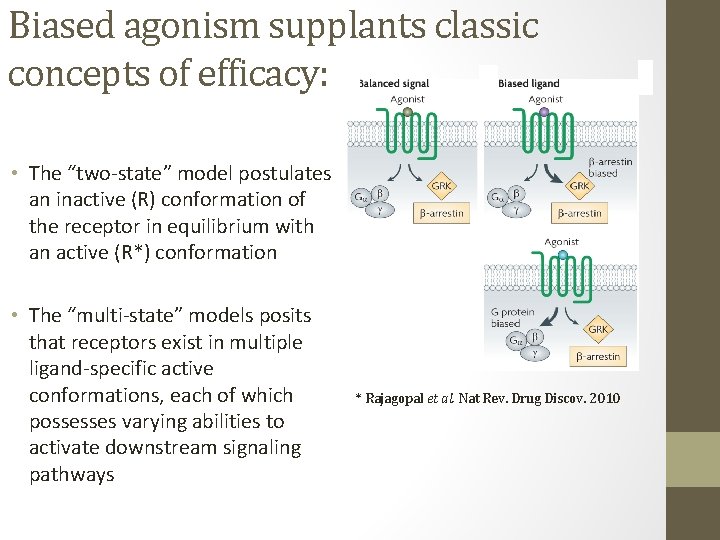
Biased agonism supplants classic concepts of efficacy: • The “two-state” model postulates an inactive (R) conformation of the receptor in equilibrium with an active (R*) conformation • The “multi-state” models posits that receptors exist in multiple ligand-specific active conformations, each of which possesses varying abilities to activate downstream signaling pathways * Rajagopal et al. Nat Rev. Drug Discov. 2010

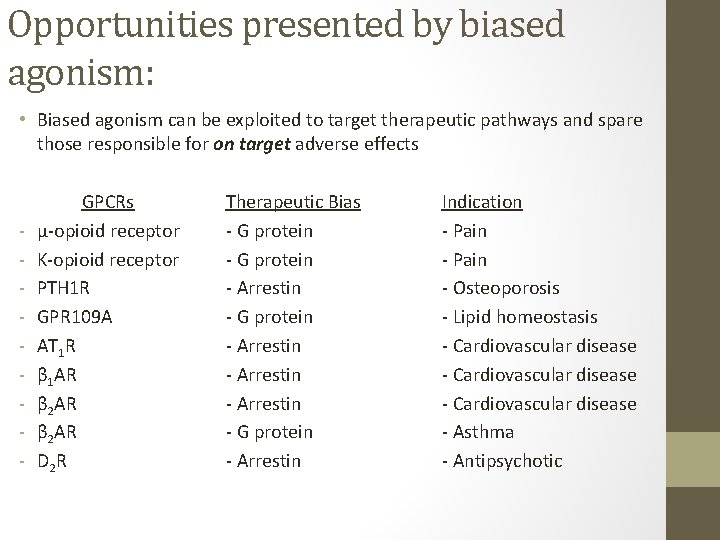
Opportunities presented by biased agonism: • Biased agonism can be exploited to target therapeutic pathways and spare those responsible for on target adverse effects - GPCRs μ-opioid receptor Κ-opioid receptor PTH 1 R GPR 109 A AT 1 R β 1 AR β 2 AR D 2 R Therapeutic Bias - G protein - Arrestin - Arrestin - G protein - Arrestin Indication - Pain - Osteoporosis - Lipid homeostasis - Cardiovascular disease - Asthma - Antipsychotic

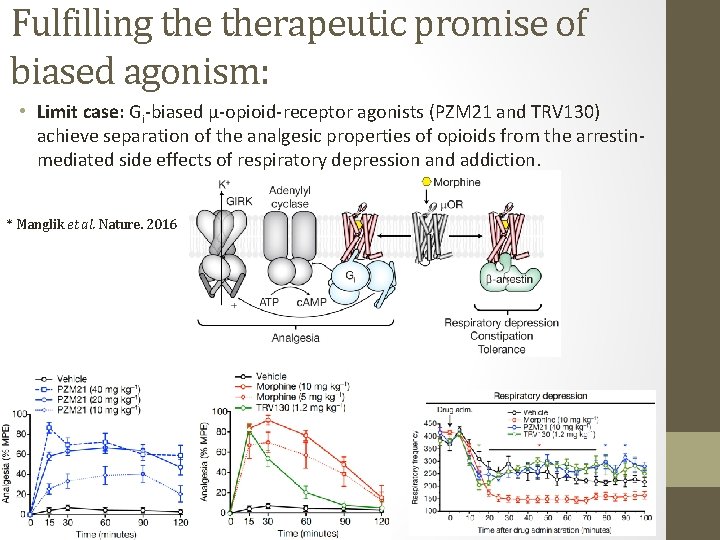
Fulfilling therapeutic promise of biased agonism: • Limit case: Gi-biased μ-opioid-receptor agonists (PZM 21 and TRV 130) achieve separation of the analgesic properties of opioids from the arrestinmediated side effects of respiratory depression and addiction. * Manglik et al. Nature. 2016

The challenges presented by biased agonism: • Hurdle: Screening for biased agonists is not straightforward and requires a reference agonist • It is difficult to extracting meaningful information about agonist efficacy from complex cellular assays with varying degrees of signal amplification • Analytical efforts to address this have been hotly debated, yet effective • Transduction coefficients (tau/KA) (Kenakin et al. ACS Chem. Neurosci. 2012) • Emax/EC 50 (equiactive comparison) (Figueroa et al. J. Pharmacol. Exp. Ther. 2009) • Tau values (pharmacologic) (Rajagopal et al. Mol. Pharmacol. 2011)

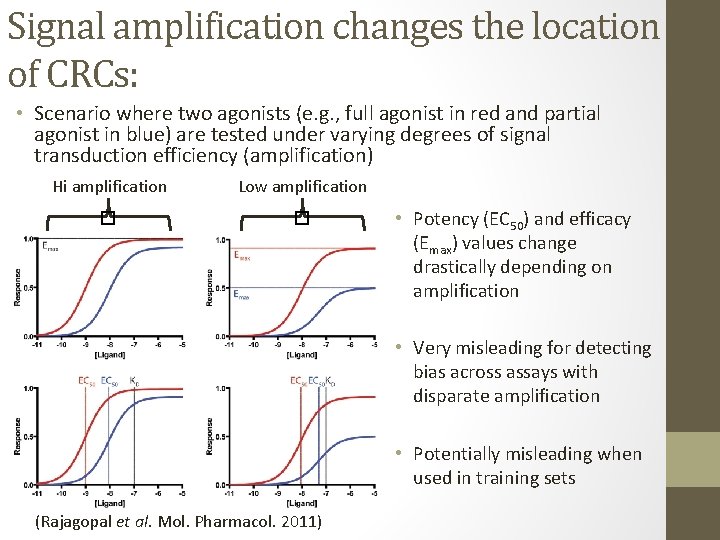
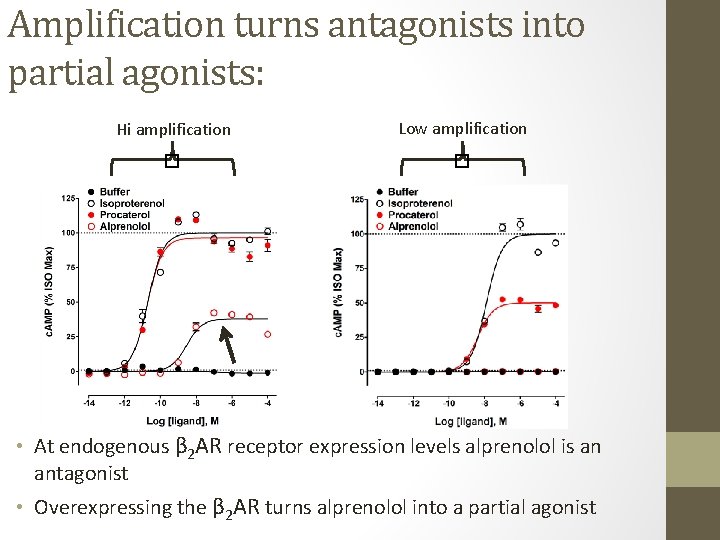
Signal amplification changes the location of CRCs: • Scenario where two agonists (e. g. , full agonist in red and partial agonist in blue) are tested under varying degrees of signal transduction efficiency (amplification) Low amplification � � Hi amplification • Potency (EC 50) and efficacy (Emax) values change drastically depending on amplification • Very misleading for detecting bias across assays with disparate amplification • Potentially misleading when used in training sets (Rajagopal et al. Mol. Pharmacol. 2011)

Hi amplification Low amplification � � Amplification turns antagonists into partial agonists: • At endogenous β 2 AR receptor expression levels alprenolol is an antagonist • Overexpressing the β 2 AR turns alprenolol into a partial agonist

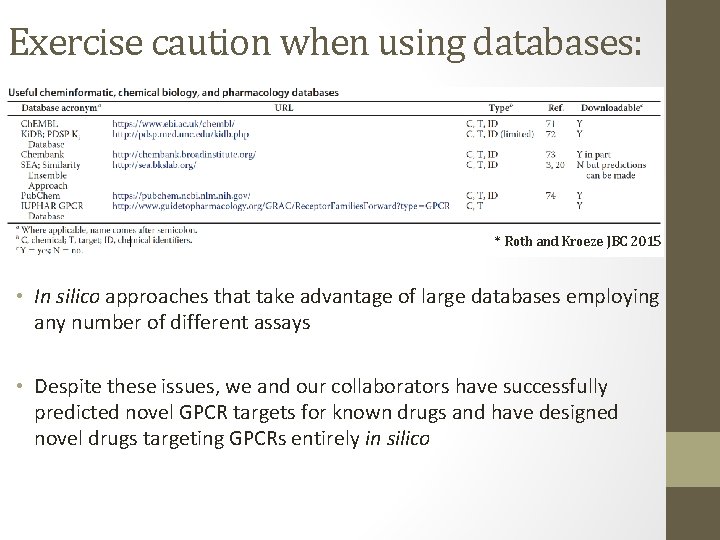
Exercise caution when using databases: * Roth and Kroeze JBC 2015 • In silico approaches that take advantage of large databases employing any number of different assays • Despite these issues, we and our collaborators have successfully predicted novel GPCR targets for known drugs and have designed novel drugs targeting GPCRs entirely in silico

If cellular assays pose such a problem, then why don’t we bypass them?

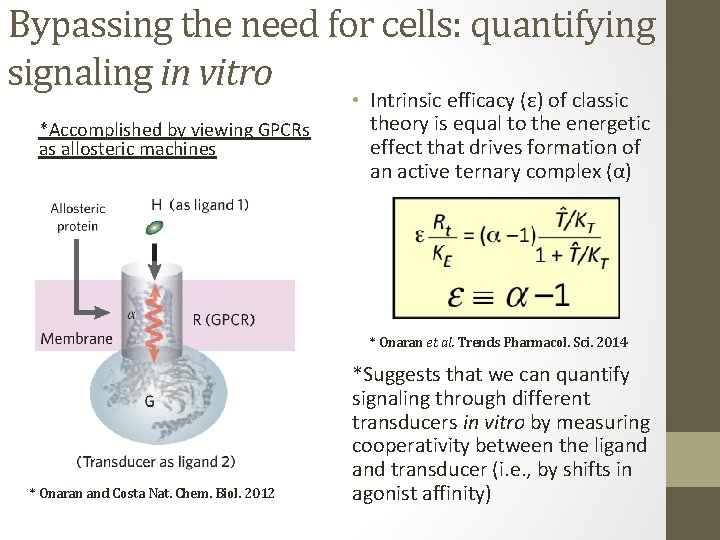
Bypassing the need for cells: quantifying signaling in vitro *Accomplished by viewing GPCRs as allosteric machines • Intrinsic efficacy (ε) of classic theory is equal to the energetic effect that drives formation of an active ternary complex (α) * Onaran et al. Trends Pharmacol. Sci. 2014 * Onaran and Costa Nat. Chem. Biol. 2012 *Suggests that we can quantify signaling through different transducers in vitro by measuring cooperativity between the ligand transducer (i. e. , by shifts in agonist affinity)

Quantifying signaling in vitro is nothing new: • Coincident with development of the Ternary Complex Model (TCM) it was shown that shifts in agonist affinity (molecular efficacy) correlate intrinsic efficacy in cells *De Lean et al. JBC. 1980 *Kent et al. Mol. Pharmacol. 1980

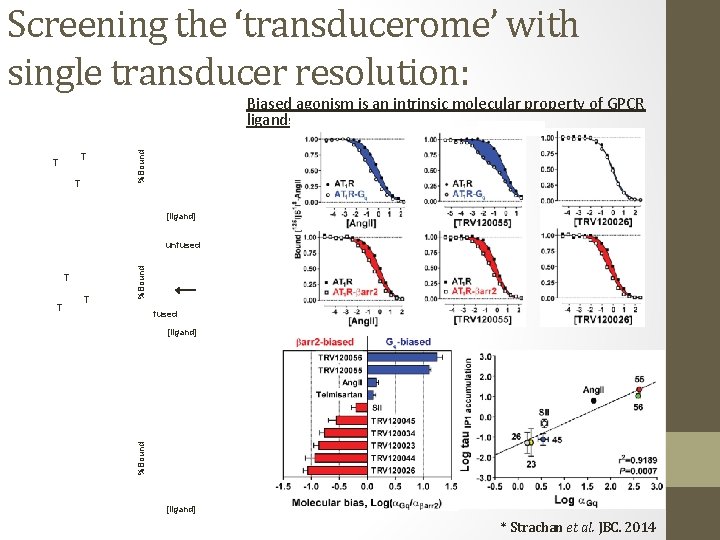
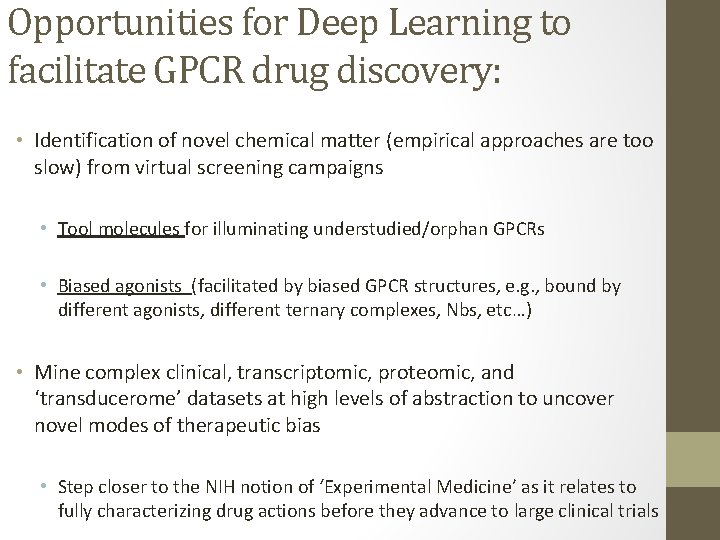
Screening the ‘transducerome’ with single transducer resolution: T T T %Bound Biased agonism is an intrinsic molecular property of GPCR ligands [ligand] T T T %Bound unfused T T T %Bound [ligand] * Strachan et al. JBC. 2014
![ligand T T T Bound unfused ligand T T Transducer This would require [ligand] T T T %Bound unfused [ligand] T T Transducer • This would require](https://slidetodoc.com/presentation_image/606e74e499ae9db3b1e7947f54d0e8fb/image-27.jpg)
[ligand] T T T %Bound unfused [ligand] T T Transducer • This would require patterns to be extracted from complex data sets • e. g. , transducerome shifts, clinical endpoints, gene expression, and behavioral data %Bound T Unfused T Area between curves T T %Bound Goal: Mine the unexplored pharmacology of GPCRs for new modes of signaling bias: [ligand] • To our knowledge no one is thinking on this scale

Summary: • The field of GPCR Molecular Pharmacology is rapidly changing, reinvigorated by paradigm shifts related to the notions that: • A large fraction of receptors are understudied or ‘orphaned’ • Biased agonism is a property of GPCR ligands • Paradigm shifts afford both numerous opportunities AND challenges • We have a long way to go in order to fully exploit this current Golden Age of Molecular Pharmacology • I am confident that advances in crystallography and computational medicinal chemistry will help to accelerate discoveries

Opportunities for Deep Learning to facilitate GPCR drug discovery: • Identification of novel chemical matter (empirical approaches are too slow) from virtual screening campaigns • Tool molecules for illuminating understudied/orphan GPCRs • Biased agonists (facilitated by biased GPCR structures, e. g. , bound by different agonists, different ternary complexes, Nbs, etc…) • Mine complex clinical, transcriptomic, proteomic, and ‘transducerome’ datasets at high levels of abstraction to uncover novel modes of therapeutic bias • Step closer to the NIH notion of ‘Experimental Medicine’ as it relates to fully characterizing drug actions before they advance to large clinical trials

The call to collaborate: successful integration of wet bench pharmacology and computation Empirical screens *Collaboration has been essential Computation/ prediction Goal: Establish a project devoted to generating the optimal AI training set for GPCR ligand discovery Target: Well-characterized GPCR family with multiple crystal structures (e. g. , opiate receptors) Ligands: Large library containing multiple chemotypes, with substantial SAR within each chemotype Data (raw and corrected): - Binding affinities (Ki’s through the Psychoactive Drug Screening Program) - Efficacy values (tau for standard assays such as Ca 2+ release, c. AMP, arrestin recruitment; use the Psychoactive Drug Screening Program ) - Molecular efficacy values from ‘transducerome screening’

Thank you! Questions?
 Orphan gpcr
Orphan gpcr Chapter summaries of lord of the flies
Chapter summaries of lord of the flies Giant molecular structure vs simple molecular structure
Giant molecular structure vs simple molecular structure Physical state of covalent compounds
Physical state of covalent compounds Ionic covalent metallic
Ionic covalent metallic Mechanism of drug action
Mechanism of drug action Pharmacology basics
Pharmacology basics Annual review of pharmacology and toxicology
Annual review of pharmacology and toxicology Essential drug concept in pharmacology
Essential drug concept in pharmacology First pass effect
First pass effect Clinical pharmacology
Clinical pharmacology Renal excretion ratio
Renal excretion ratio Pharmacology module
Pharmacology module Dose adjustment in renal and hepatic disease
Dose adjustment in renal and hepatic disease Adrenal drugs pharmacology
Adrenal drugs pharmacology Slidetodoc
Slidetodoc Estado estable
Estado estable Pharmacology for veterinary technicians
Pharmacology for veterinary technicians First bypass effect
First bypass effect Rationale meaning in pharmacology
Rationale meaning in pharmacology If time permits quotes
If time permits quotes Chapter 15 diagnostic procedures and pharmacology
Chapter 15 diagnostic procedures and pharmacology First pass effect in pharmacology
First pass effect in pharmacology Fish pharmacology
Fish pharmacology Dopamine pharmacology
Dopamine pharmacology Samyukti meaning
Samyukti meaning Basic & clinical pharmacology
Basic & clinical pharmacology Hepatic extraction ratio formula
Hepatic extraction ratio formula What is ion trapping in pharmacology
What is ion trapping in pharmacology Pharmacology pay
Pharmacology pay Pharmacology introduction
Pharmacology introduction Mdi pharmacology
Mdi pharmacology