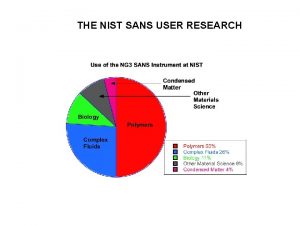
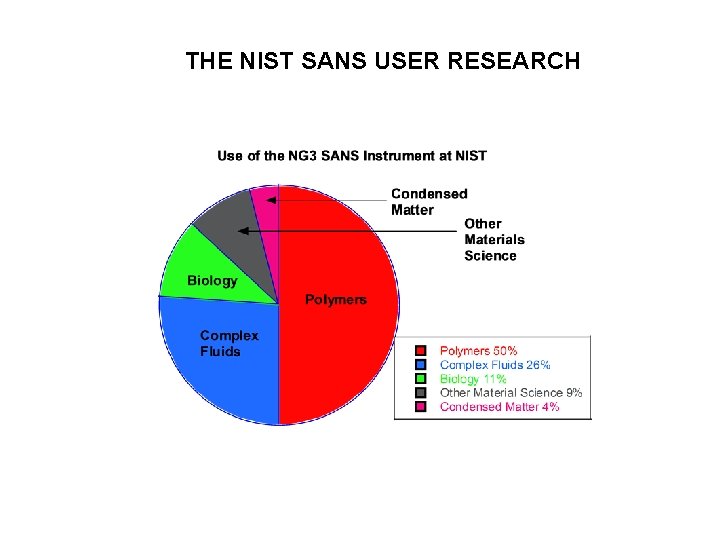
THE NIST SANS USER RESEARCH SANS RESEARCH TOPICS




![NONLINEAR LEAST-SQUARES FIT Functional form: I(Q) = C/[1+(QL)m] + Background C: solvation intensity L: NONLINEAR LEAST-SQUARES FIT Functional form: I(Q) = C/[1+(QL)m] + Background C: solvation intensity L:](https://slidetodoc.com/presentation_image_h2/0ee5453d5e6b16ee91c98eba3b4fdb1d/image-24.jpg)

- Slides: 29

THE NIST SANS USER RESEARCH

SANS RESEARCH TOPICS Boualem Hammouda National Institute of Standards and Technology Center for Neutron Research 1. SANS from Pluronics 2. Polymer Blend Thermodynamics 3. Helix-to-Coil Transition in DNA

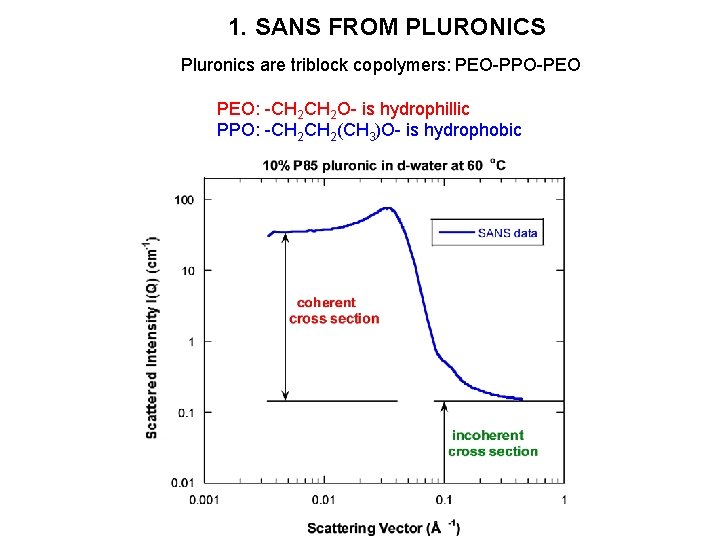
1. SANS FROM PLURONICS Pluronics are triblock copolymers: PEO-PPO-PEO PEO: -CH 2 O- is hydrophillic PPO: -CH 2(CH 3)O- is hydrophobic

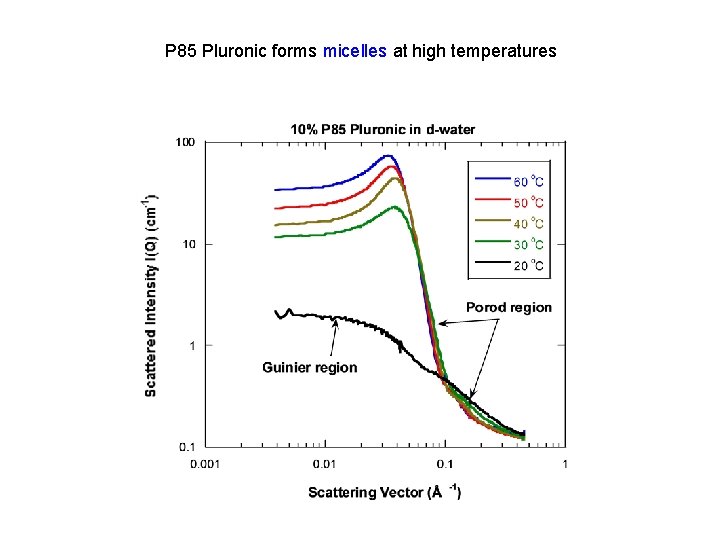
P 85 Pluronic forms micelles at high temperatures

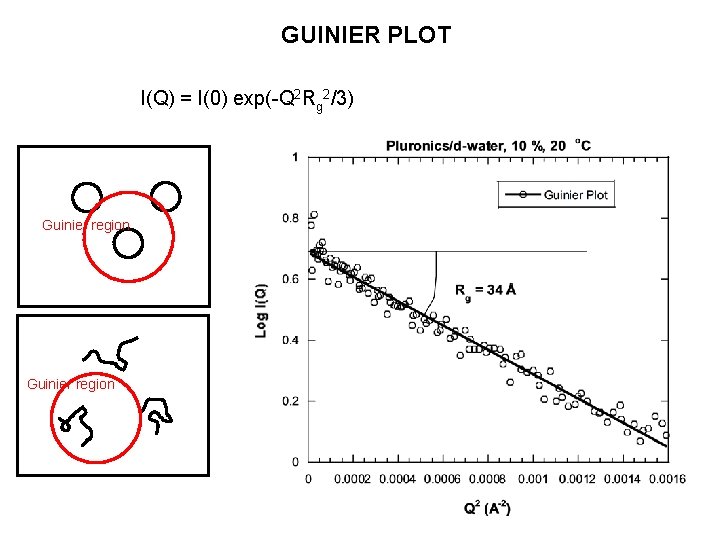
GUINIER PLOT I(Q) = I(0) exp(-Q 2 Rg 2/3) Guinier region

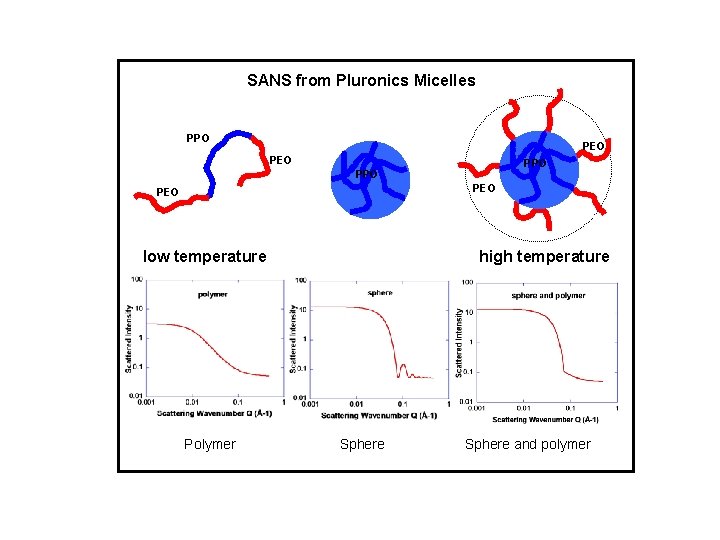
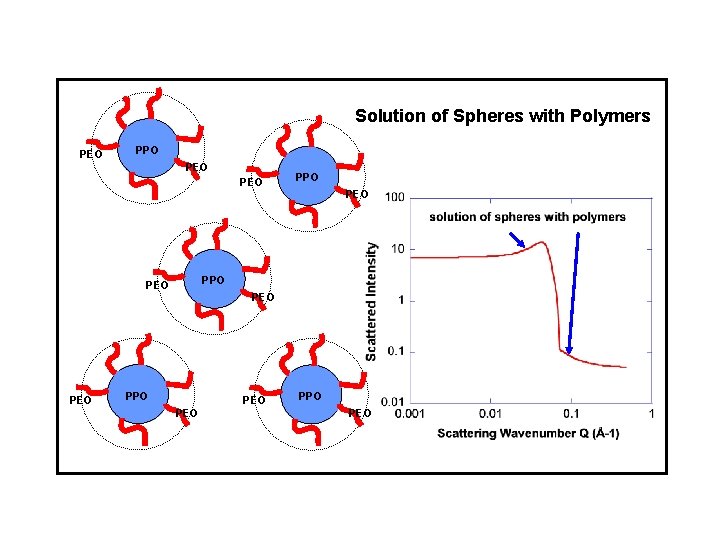
SANS from Pluronics Micelles PPO PEO low temperature Polymer high temperature Sphere and polymer

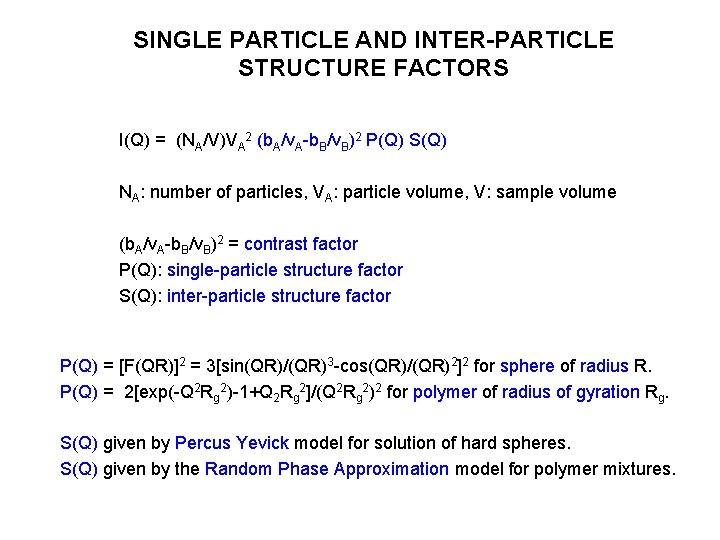
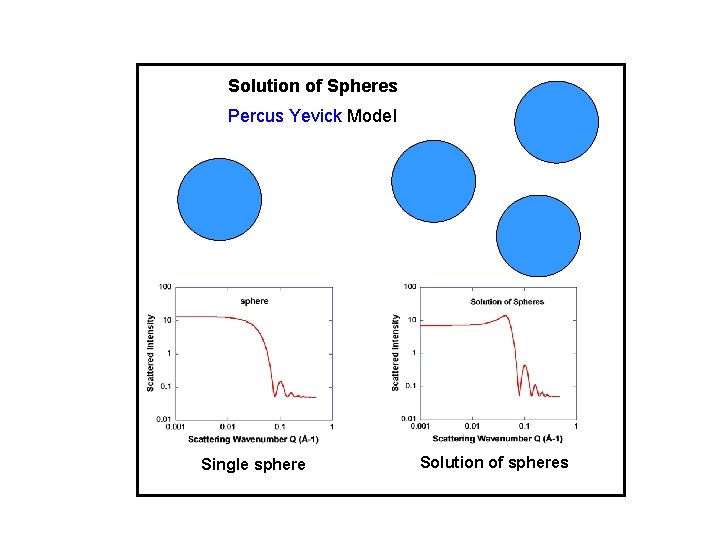
SINGLE PARTICLE AND INTER-PARTICLE STRUCTURE FACTORS I(Q) = (NA/V)VA 2 (b. A/v. A-b. B/v. B)2 P(Q) S(Q) NA: number of particles, VA: particle volume, V: sample volume (b. A/v. A-b. B/v. B)2 = contrast factor P(Q): single-particle structure factor S(Q): inter-particle structure factor P(Q) = [F(QR)]2 = 3[sin(QR)/(QR)3 -cos(QR)/(QR)2]2 for sphere of radius R. P(Q) = 2[exp(-Q 2 Rg 2)-1+Q 2 Rg 2]/(Q 2 Rg 2)2 for polymer of radius of gyration Rg. S(Q) given by Percus Yevick model for solution of hard spheres. S(Q) given by the Random Phase Approximation model for polymer mixtures.

Solution of Spheres Percus Yevick Model Single sphere Solution of spheres

Solution of Spheres with Polymers PEO PPO PEO PEO PPO PEO

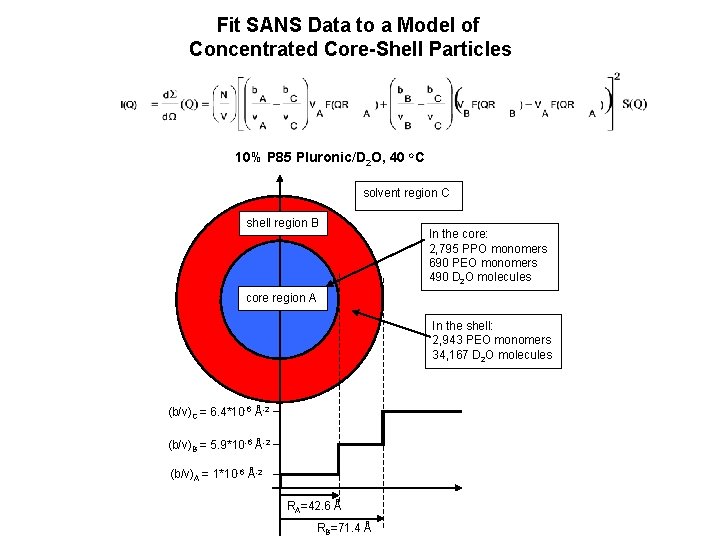
Fit SANS Data to a Model of Concentrated Core-Shell Particles 10% P 85 Pluronic/D 2 O, 40 o. C solvent region C shell region B In the core: 2, 795 PPO monomers 690 PEO monomers 490 D 2 O molecules core region A In the shell: 2, 943 PEO monomers 34, 167 D 2 O molecules (b/v)C = 6. 4*10 -6 Å-2 (b/v)B = 5. 9*10 -6 Å-2 (b/v)A = 1*10 -6 Å-2 RA=42. 6 Å RB=71. 4 Å

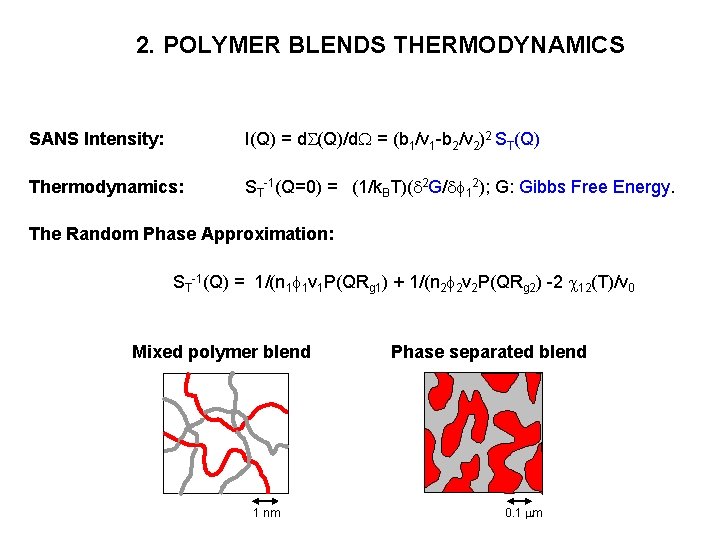
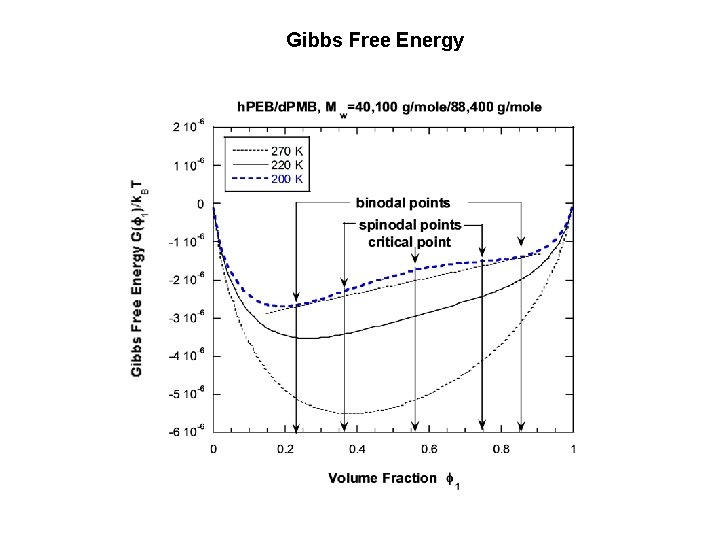
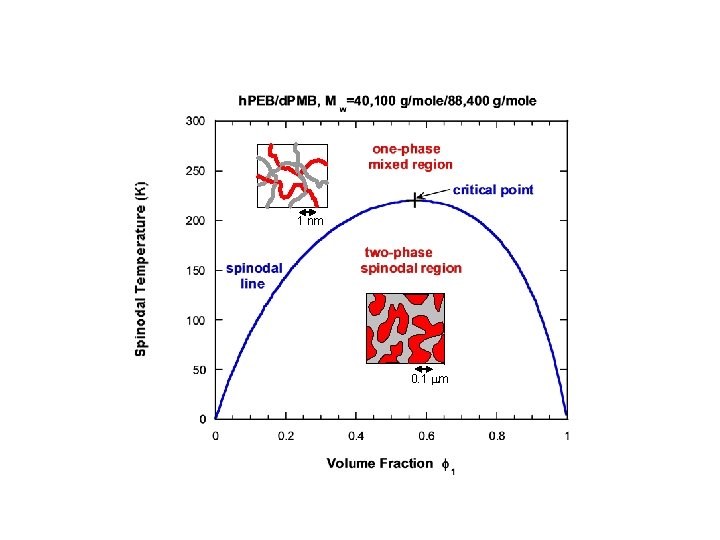
2. POLYMER BLENDS THERMODYNAMICS SANS Intensity: I(Q) = d. S(Q)/d. W = (b 1/v 1 -b 2/v 2)2 ST(Q) Thermodynamics: ST-1(Q=0) = (1/k. BT)(d 2 G/df 12); G: Gibbs Free Energy. The Random Phase Approximation: ST-1(Q) = 1/(n 1 f 1 v 1 P(QRg 1) + 1/(n 2 f 2 v 2 P(QRg 2) -2 c 12(T)/v 0 Mixed polymer blend 1 nm Phase separated blend 0. 1 mm

Gibbs Free Energy


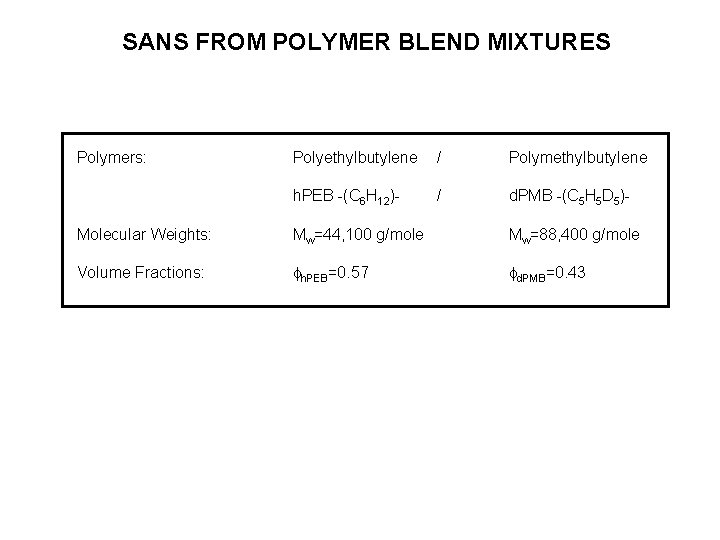
SANS FROM POLYMER BLEND MIXTURES Polymers: Polyethylbutylene / Polymethylbutylene h. PEB -(C 6 H 12)- / d. PMB -(C 5 H 5 D 5)- Molecular Weights: Mw=44, 100 g/mole Mw=88, 400 g/mole Volume Fractions: fh. PEB=0. 57 fd. PMB=0. 43


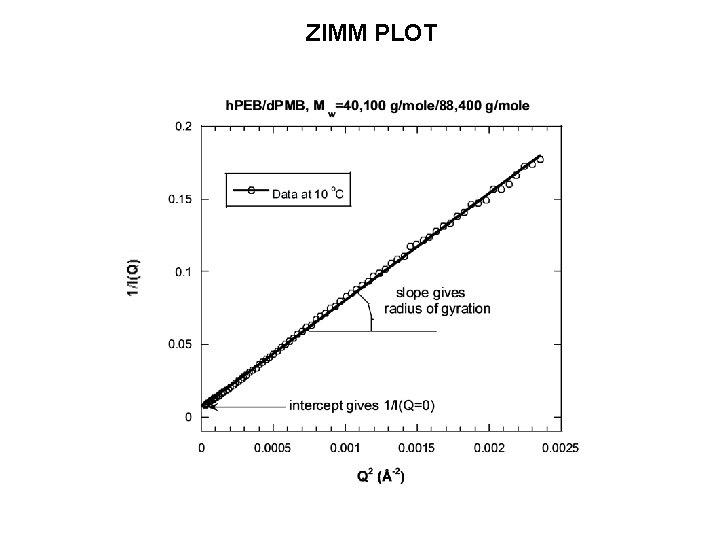
ZIMM PLOT


1 nm 0. 1 mm

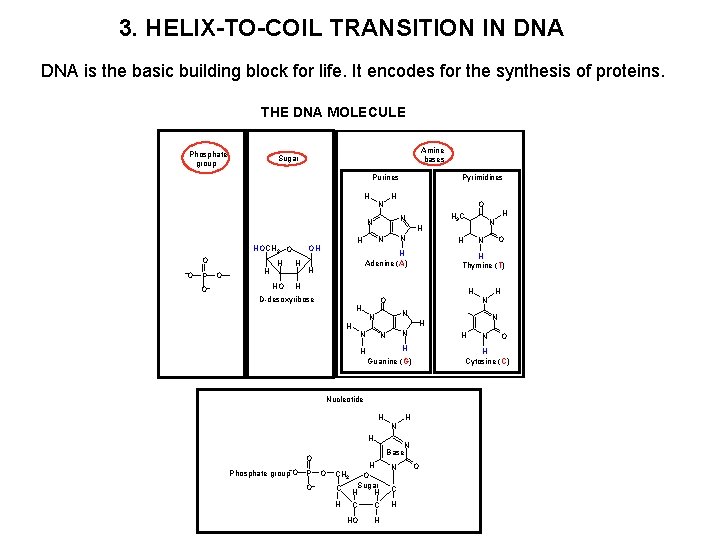
3. HELIX-TO-COIL TRANSITION IN DNA is the basic building block for life. It encodes for the synthesis of proteins. THE DNA MOLECULE Phosphate group Amine bases Sugar Purines H N H O O P O O H HO H N N H H Adenine (A) H D-desoxyribose N N H H Nucleotide H Phosphate group O P O O N H N Base N C H O Sugar H H H C C H HO H CH 2 C N O H Cytosine (C) Guanine (G) H H N H H O N N N O H Thymine (T) H N O H H N H H OH O H 3 C N N HOCH 2 O Pyrimidines O

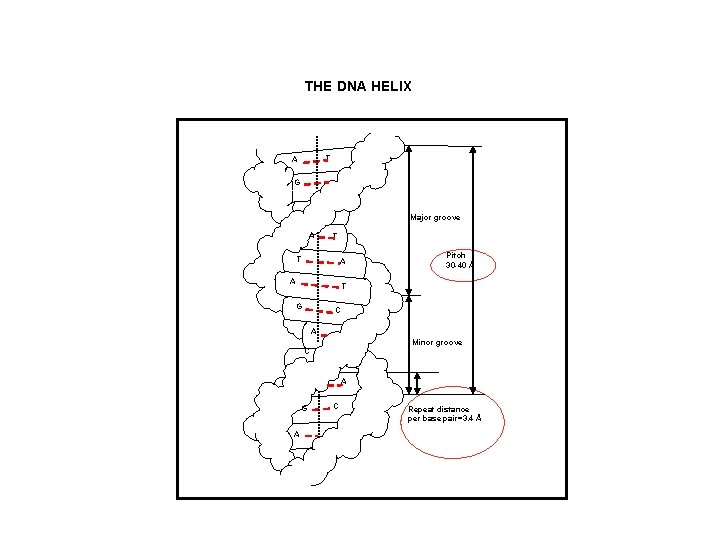
THE DNA HELIX T A G Major groove A T T A A Pitch 30 -40 Å T G C A Minor groove C A G A C Repeat distance per base pair=3. 4 Å

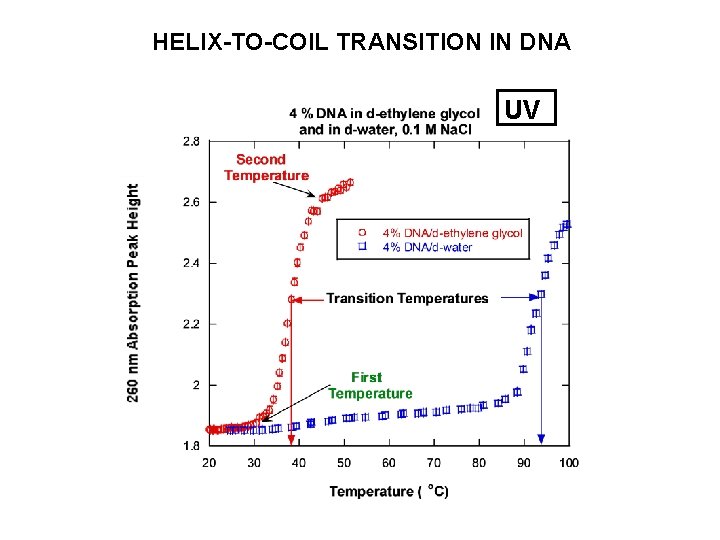
HELIX-TO-COIL TRANSITION IN DNA UV

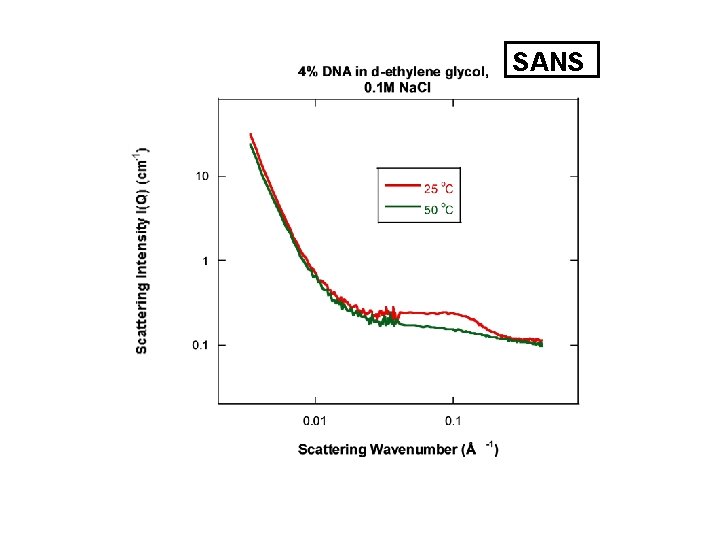
SANS

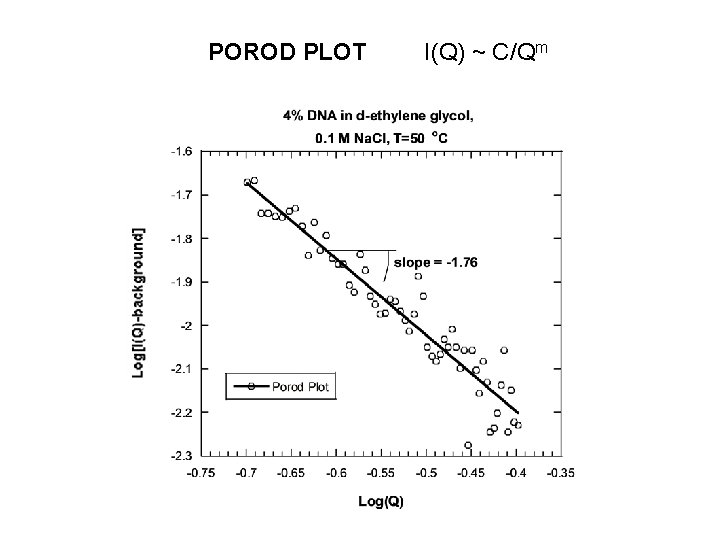
POROD PLOT I(Q) ~ C/Qm
![NONLINEAR LEASTSQUARES FIT Functional form IQ C1QLm Background C solvation intensity L NONLINEAR LEAST-SQUARES FIT Functional form: I(Q) = C/[1+(QL)m] + Background C: solvation intensity L:](https://slidetodoc.com/presentation_image_h2/0ee5453d5e6b16ee91c98eba3b4fdb1d/image-24.jpg)
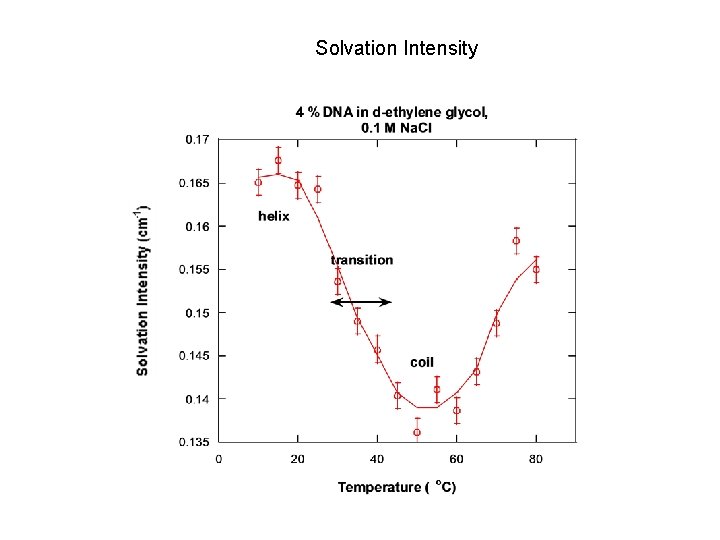
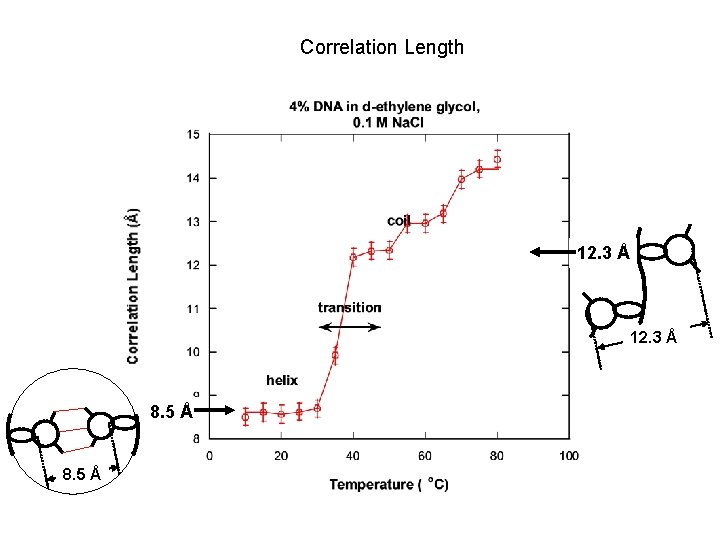
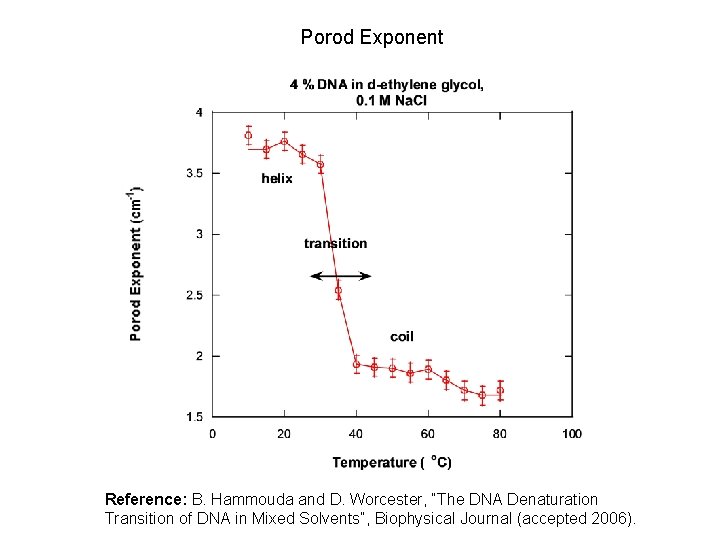
NONLINEAR LEAST-SQUARES FIT Functional form: I(Q) = C/[1+(QL)m] + Background C: solvation intensity L: correlation length m: Porod exponent

Solvation Intensity

Correlation Length 12. 3 Å 8. 5 Å

Porod Exponent Reference: B. Hammouda and D. Worcester, “The DNA Denaturation Transition of DNA in Mixed Solvents”, Biophysical Journal (accepted 2006).

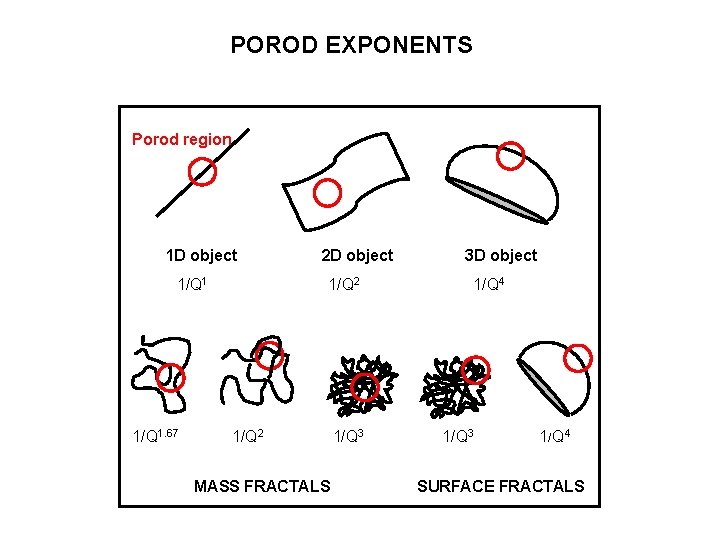
POROD EXPONENTS Porod region 1 D object 1/Q 1. 67 2 D object 1/Q 2 MASS FRACTALS 1/Q 3 3 D object 1/Q 4 1/Q 3 1/Q 4 SURFACE FRACTALS

CONCLUSIONS -- The SANS technique is a valuable characterization method. -- SANS has been effective in complex fluids, polymers, biology, etc. -- SANS can determine structures, phase transitions, and morphology. -- The NG 3 SANS instrument at NIST gets over 150 users per year, resulting in over 40 publications per year. ACKNOWLEDGMENTS NSF-DMR, Steve Kline, Nitash Balsara, David Worcester. CHECK IT OUT: http: //www. ncnr. nist. gov/programs/sans/ http: //www. ncnr. nist. gov/staff/hammouda/ hammouda@nist. gov
 Ont peut vivre sans richesse
Ont peut vivre sans richesse Qu'est-ce que tu aimes manger?
Qu'est-ce que tu aimes manger? Single user and multiple user operating system
Single user and multiple user operating system Single user and multi user operating system
Single user and multi user operating system Operations research topics
Operations research topics Research topics and their objectives
Research topics and their objectives Global question examples
Global question examples Technology research topics
Technology research topics Tahapan penelitian dalam psikologi
Tahapan penelitian dalam psikologi Wireless communication research topics
Wireless communication research topics Efl research topics
Efl research topics Action research topics in physics
Action research topics in physics Mobile computing research topics
Mobile computing research topics Data warehouse research topics
Data warehouse research topics Philosophical assumption
Philosophical assumption Content analysis research topics
Content analysis research topics Pob sba recommendation sample
Pob sba recommendation sample Business cognate sba
Business cognate sba Non experimental research design
Non experimental research design Operations research topics
Operations research topics Observation in research
Observation in research Hình ảnh bộ gõ cơ thể búng tay
Hình ảnh bộ gõ cơ thể búng tay Lp html
Lp html Bổ thể
Bổ thể Tỉ lệ cơ thể trẻ em
Tỉ lệ cơ thể trẻ em Gấu đi như thế nào
Gấu đi như thế nào Glasgow thang điểm
Glasgow thang điểm Hát lên người ơi
Hát lên người ơi Kể tên các môn thể thao
Kể tên các môn thể thao Thế nào là hệ số cao nhất
Thế nào là hệ số cao nhất