The Cancer genome atlas TCGA and the search




- Slides: 15

The Cancer genome atlas (TCGA) and the search for a CUP genetic/epigenetic signature Manel Esteller, MD, Ph. D. Director, Josep Carreras Leukaemia Research Institute (IJC) Director, Cancer Epigenetics and Biology Program (PEBC) ICREA Research Professor Genetics Chairman, School of Medicine, University of Barcelona

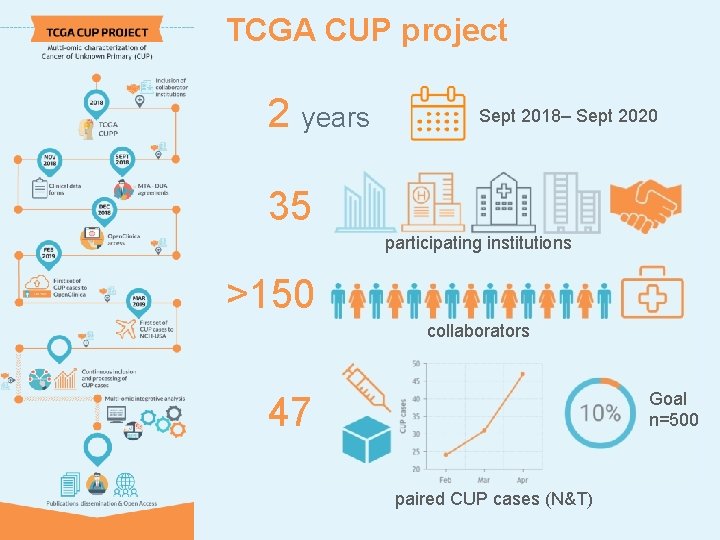
TCGA CUP project 2 years Sept 2018– Sept 2020 35 participating institutions >150 collaborators Goal n=500 47 paired CUP cases (N&T)

Project budget Cost to NCI is approximately: Tissue Acquisition: $500 per case, for a total of $250, 000 Tissue Processing: $2000/per case, for a total cost of $1, 000. Tissue Characterization: $5000/case, for a total of 2, 500, 000 Total Investment: $3, 750, 000$

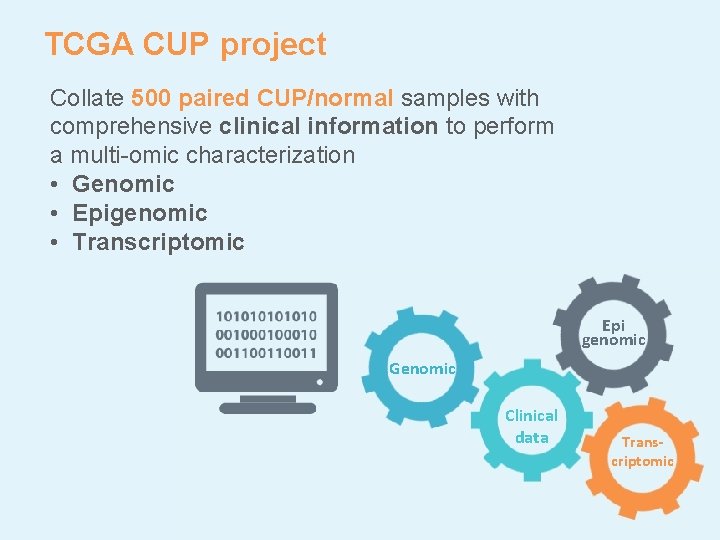
TCGA CUP project Collate 500 paired CUP/normal samples with comprehensive clinical information to perform a multi-omic characterization • Genomic • Epigenomic • Transcriptomic Epi genomic Genomic Clinical data Transcriptomic

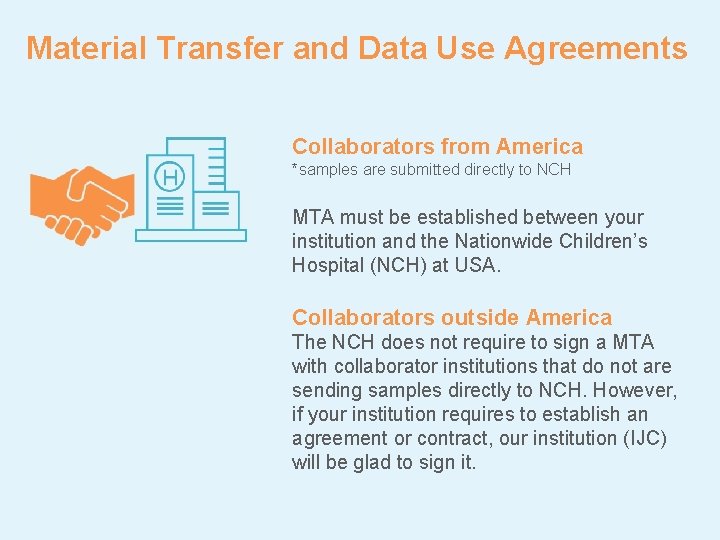
Material Transfer and Data Use Agreements Collaborators from America *samples are submitted directly to NCH MTA must be established between your institution and the Nationwide Children’s Hospital (NCH) at USA. Collaborators outside America The NCH does not require to sign a MTA with collaborator institutions that do not are sending samples directly to NCH. However, if your institution requires to establish an agreement or contract, our institution (IJC) will be glad to sign it.

TGCA CUPP collaborator institutions 1 2 3 4 15

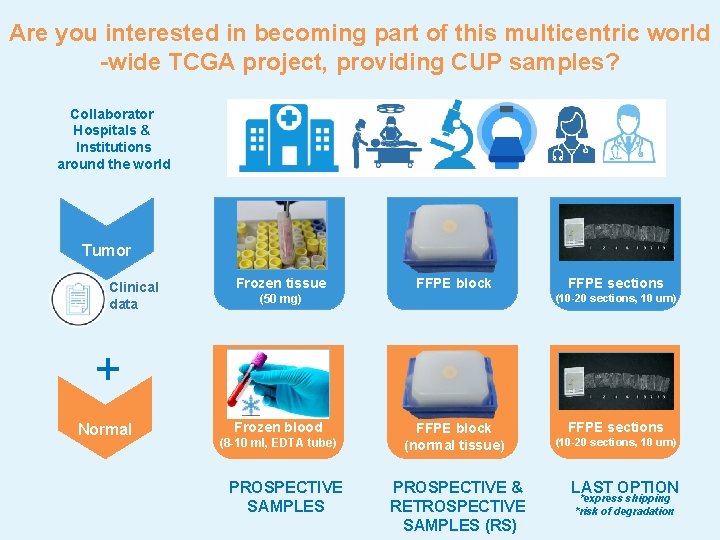
Are you interested in becoming part of this multicentric world -wide TCGA project, providing CUP samples? Collaborator Hospitals & Institutions around the world Tumor Clinical data Frozen tissue FFPE block FFPE sections (10 -20 sections, 10 um) (50 mg) + Normal Frozen blood (8 -10 ml, EDTA tube) PROSPECTIVE SAMPLES FFPE block (normal tissue) PROSPECTIVE & RETROSPECTIVE SAMPLES (RS) FFPE sections (10 -20 sections, 10 um) LAST OPTION *express shipping *risk of degradation

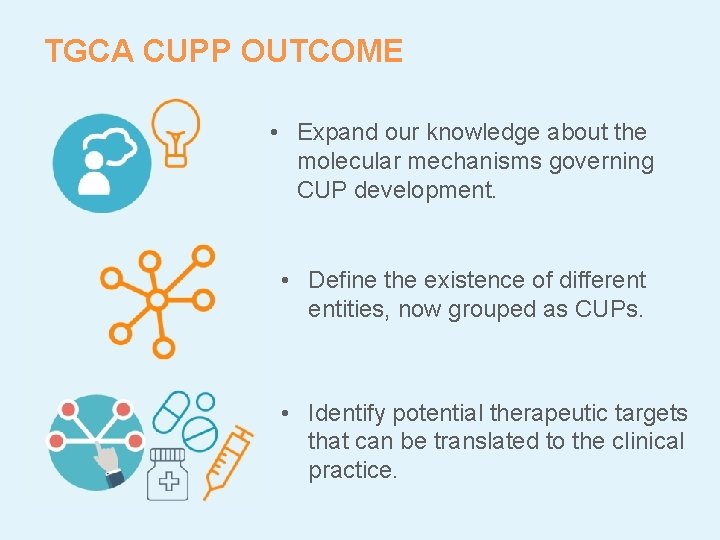
TGCA CUPP OUTCOME • Expand our knowledge about the molecular mechanisms governing CUP development. • Define the existence of different entities, now grouped as CUPs. • Identify potential therapeutic targets that can be translated to the clinical practice.

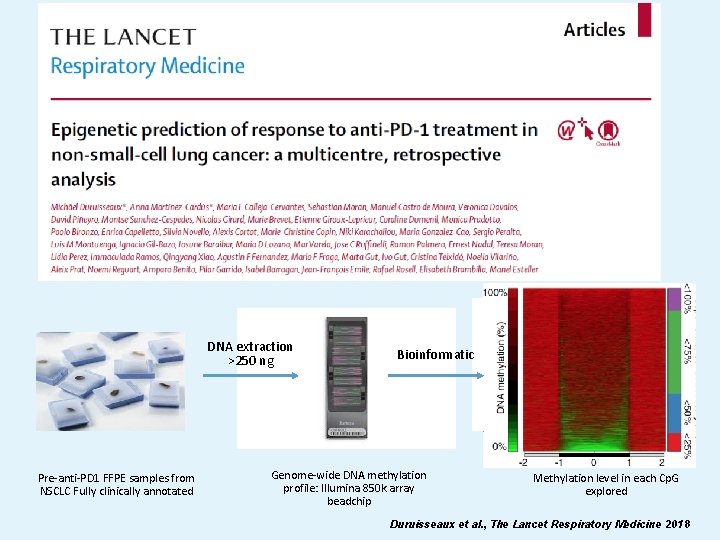
DNA extraction >250 ng Pre-anti-PD 1 FFPE samples from NSCLC Fully clinically annotated Bioinformatic Genome-wide DNA methylation profile: Illumina 850 k array beadchip Methylation level in each Cp. G explored Duruisseaux et al. , The Lancet Respiratory Medicine 2018

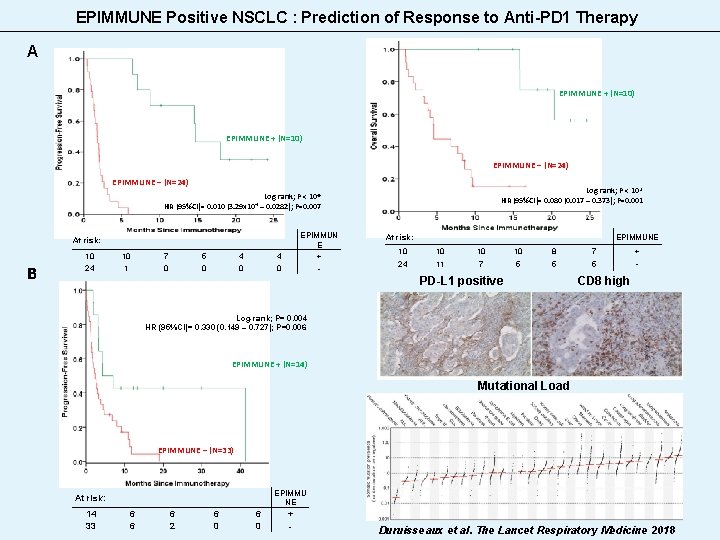
EPIMMUNE Positive NSCLC : Prediction of Response to Anti-PD 1 Therapy A EPIMMUNE + (N=10) EPIMMUNE – (N=24) Log-rank; P < 10 -3 HR (95%CI)= 0. 080 (0. 017 – 0. 373); P=0. 001 Log-rank; P < 10 -6 HR (95%CI)= 0. 010 (3. 29 x 10 -4 – 0. 0282); P=0. 007 10 7 5 4 4 EPIMMUN E + 24 1 0 0 - At risk: B At risk: 10 8 7 EPIMMUNE 10 24 11 7 5 5 5 - PD-L 1 positive + CD 8 high Log-rank; P= 0. 004 HR (95%CI)= 0. 330 (0. 149 – 0. 727); P=0. 006 EPIMMUNE + (N=14) Mutational Load EPIMMUNE – (N=33) At risk: 14 33 6 6 6 2 6 0 EPIMMU NE + - Duruisseaux et al. The Lancet Respiratory Medicine 2018

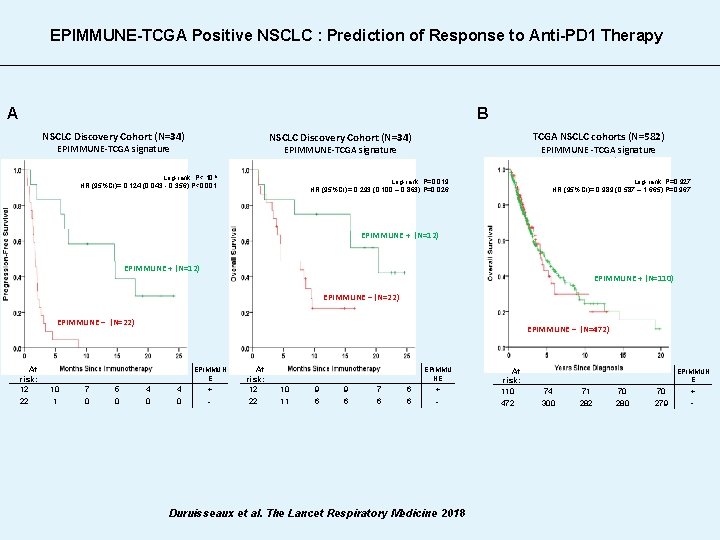
EPIMMUNE-TCGA Positive NSCLC : Prediction of Response to Anti-PD 1 Therapy A B NSCLC Discovery Cohort (N=34) TCGA NSCLC cohorts (N=582) NSCLC Discovery Cohort (N=34) EPIMMUNE-TCGA signature EPIMMUNE-TCGA signature Log-rank; P< 10 -5 HR (95%CI)= 0. 124 (0. 043 - 0. 356); P<0. 001 Log-rank; P=0. 927 HR (95%CI)= 0. 989 (0. 587 – 1. 665); P=0. 967 Log-rank; P=0. 019 HR (95%CI)= 0. 293 (0. 100 – 0. 863); P=0. 026 EPIMMUNE + (N=12) EPIMMUNE + (N=110) EPIMMUNE – (N=22) At risk: 12 22 10 1 7 0 5 0 EPIMMUNE – (N=472) 4 0 EPIMMUN E + - At risk: 12 22 10 11 9 6 7 6 6 6 EPIMMU NE + - Duruisseaux et al. The Lancet Respiratory Medicine 2018 At risk: 110 472 74 300 71 282 70 280 70 279 EPIMMUN E + -

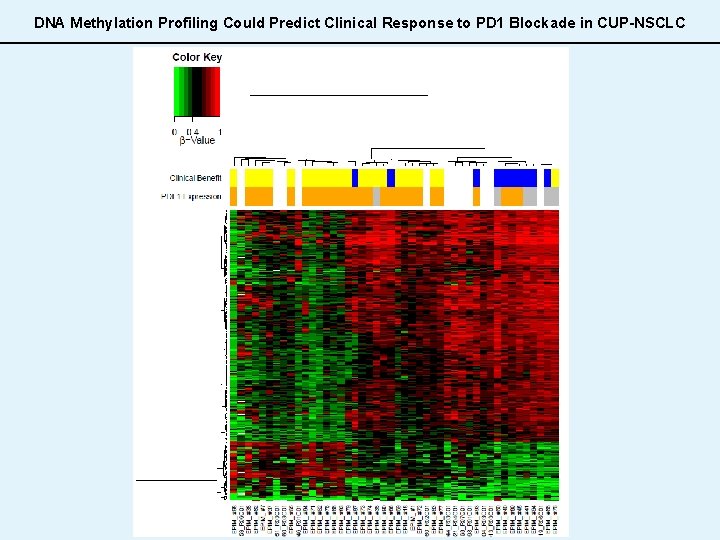
DNA Methylation Profiling Could Predict Clinical Response to PD 1 Blockade in CUP-NSCLC

BENEFITS FOR COLLABORATORS • Access to multi-omic data for CUP cases. • Authorship in top high-impact publications (i. e. Nature, Science, Cell…)

mesteller@carrerasresearch. org

 Semi-global alignment
Semi-global alignment Bioportal tcga
Bioportal tcga Biologists search the volumes of the human genome using
Biologists search the volumes of the human genome using De novo genome assembly ppt
De novo genome assembly ppt Informed and uninformed search
Informed and uninformed search Genome
Genome Plant genome research program
Plant genome research program Euphenics
Euphenics Stanford
Stanford Human genome size
Human genome size Mash distance
Mash distance Human genome size
Human genome size Difference between bac and yac
Difference between bac and yac Satellite dna
Satellite dna Perpartes
Perpartes Human genome structure
Human genome structure