The Cancer Genome Atlas Project January 24 2008


- Slides: 10

The Cancer Genome Atlas Project January 24, 2008 TCGA

Program • Goal: find genomic alterations that cause cancer (mutations, CNA, methylation, …) • Pilot project – $100 M (NCI/NHGRI) – 3 years – 3 diseases • brain (glioblastoma multiforme) • lung (squamous) • ovarian (serous cystadenocarcinoma ) TCGA

Organization • Biospecimen Core Resource (BCR) • Genome Sequencing Centers (GSCs) (3) • Cancer Genome Characterization Centers (CGCCs) (7) • Data Coordinating Center (DCC) • Project Team (NCI/NHGRI) • Steering Committee (NCI/NHGRI & PIs) • External Scientific Committee • Working Groups TCGA

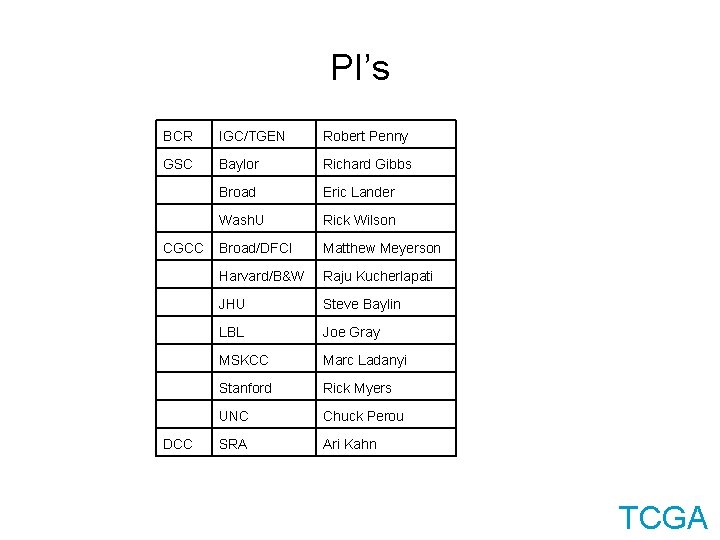
PI’s BCR IGC/TGEN Robert Penny GSC Baylor Richard Gibbs Broad Eric Lander Wash. U Rick Wilson CGCC Broad/DFCI Matthew Meyerson Harvard/B&W Raju Kucherlapati JHU Steve Baylin LBL Joe Gray MSKCC Marc Ladanyi Stanford Rick Myers UNC Chuck Perou DCC SRA Ari Kahn TCGA

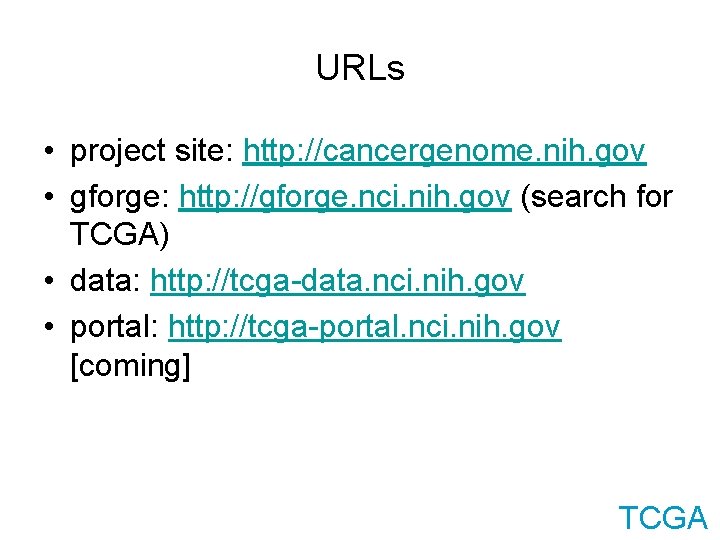
URLs • project site: http: //cancergenome. nih. gov • gforge: http: //gforge. nci. nih. gov (search for TCGA) • data: http: //tcga-data. nci. nih. gov • portal: http: //tcga-portal. nci. nih. gov [coming] TCGA

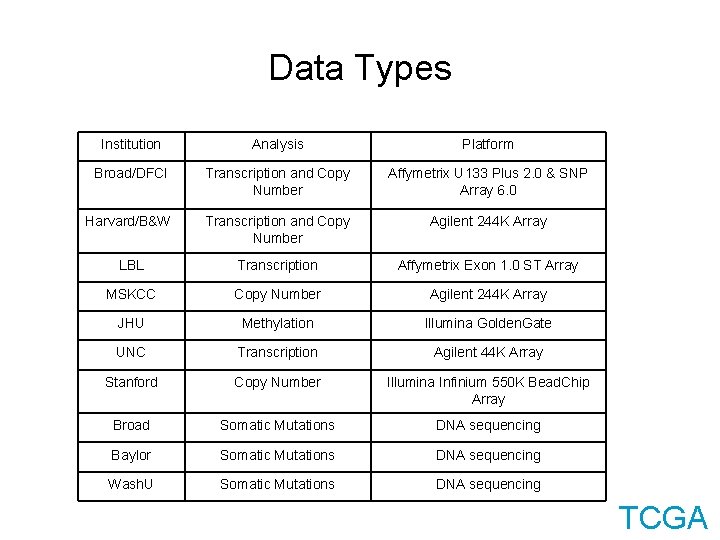
Data Types Institution Analysis Platform Broad/DFCI Transcription and Copy Number Affymetrix U 133 Plus 2. 0 & SNP Array 6. 0 Harvard/B&W Transcription and Copy Number Agilent 244 K Array LBL Transcription Affymetrix Exon 1. 0 ST Array MSKCC Copy Number Agilent 244 K Array JHU Methylation Illumina Golden. Gate UNC Transcription Agilent 44 K Array Stanford Copy Number Illumina Infinium 550 K Bead. Chip Array Broad Somatic Mutations DNA sequencing Baylor Somatic Mutations DNA sequencing Wash. U Somatic Mutations DNA sequencing TCGA

Data Levels • raw – low-level data for a single sample, not normalized (e. g. , trace file, . cel file) • processed – single-sample, normalized & interpreted (e. g. mutation call, amplification call for a locus, . snp, . chp) • segmented (n/a for mutation & expression) – single-sample, aggregation of loci into regions (e. g. amplification call for a region of a sample) • summary finding (aka “region of interest”) – cross-sample findings (e. g. minimal common region of amplification across a sample set) TCGA

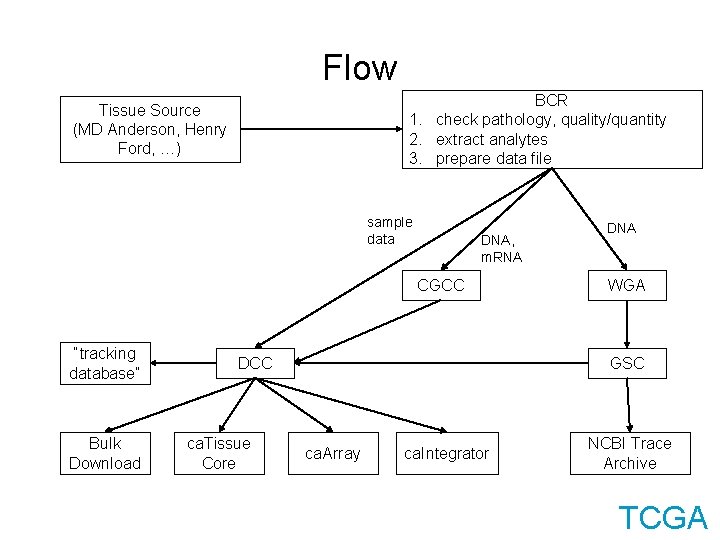
Flow BCR 1. check pathology, quality/quantity 2. extract analytes 3. prepare data file Tissue Source (MD Anderson, Henry Ford, …) sample data DNA, m. RNA CGCC “tracking database” Bulk Download DCC ca. Tissue Core DNA WGA GSC ca. Array ca. Integrator NCBI Trace Archive TCGA

Data Formats • BCR – XML (tags are CDEs) – images • GSC – Called mutations (Genboree LFF format) – Linking table • sample-trace-target • CGCC – MAGE-TAB • IDF: Investigation Definition Format • SDRF: Sample and Data Relationship Format TCGA

Where Does/Will the Data Go? • • • ftp site (now with a simple web wrapper: “portal #1”) “tracking database” repositories with ca. BIG API’s – – • • ca. Array ca. Tissue CORE ca. Integrator NCIA NCBI trace archive a richer, “portal #2” – – – – more convenient download capability filtering datasets by clinical information summary level data genome browser view gene info page visualization on pathways etc. TCGA