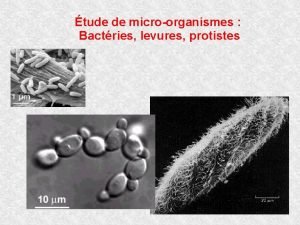
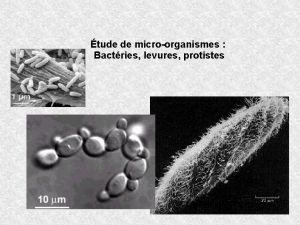
Rarrangements chromosomiques Evolution des gnomes de levures 2me




- Slides: 42

Réarrangements chromosomiques Evolution des génomes de levures 2ème réunion GTGC Nantes 12 et 13 octobre 2006 Gilles Fischer

Comparative genomics in yeasts: significant results: - Gene identification and annotation - Sequence comparison and protein evolution

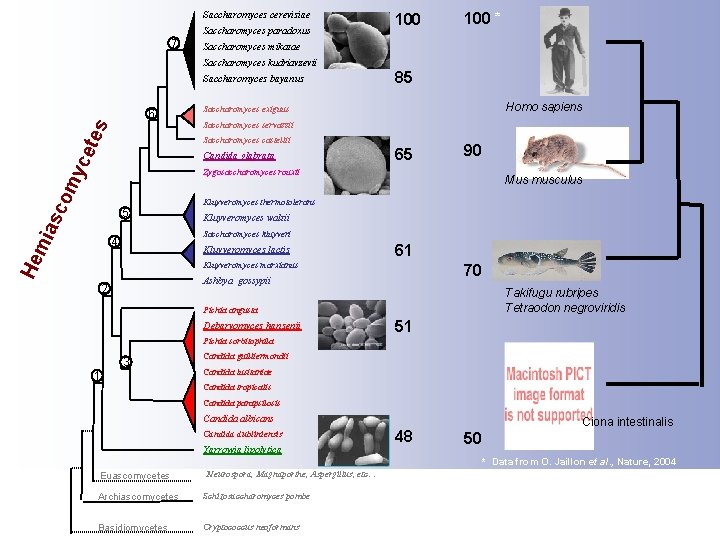
Saccharomyces cerevisiae Saccharomyces paradoxus 7 100 * Saccharomyces mikatae Saccharomyces kudriavzevii Saccharomyces bayanus tes 6 Homo sapiens Saccharomyces exiguus Saccharomyces servazzii Saccharomyces castellii yce 85 Candida glabrata 65 90 asc om Zygosaccharomyces rouxii 5 Kluyveromyces thermotolerans Kluyveromyces waltii Saccharomyces kluyveri 4 Kluyveromyces lactis He mi Mus musculus 61 Kluyveromyces marxianus 70 Ashbya gossypii 2 Takifugu rubripes Tetraodon negroviridis Pichia angusta Debaryomyces hansenii 51 Pichia sorbitophila 3 1 Candida guilliermondii Candida lusitaniae Candida tropicalis Candida parapsilosis Candida albicans Candida dubliniensis Yarrowia lipolytica 48 Ciona intestinalis 50 * Data from O. Jaillon et al. , Nature, 2004 Euascomycetes Neurospora, Magnaporthe, Aspergillus, etc… Archiascomycetes Schizosaccharomyces pombe Basidiomycetes Cryptococcus neoformans

Comparative genomics in yeasts: significant results: - Gene identification and annotation - Sequence comparison and protein evolution - Chromosome rearrangements 1> speciation process 2> level of chromosome reorganisation 3> rates of chromosomal rearrangements

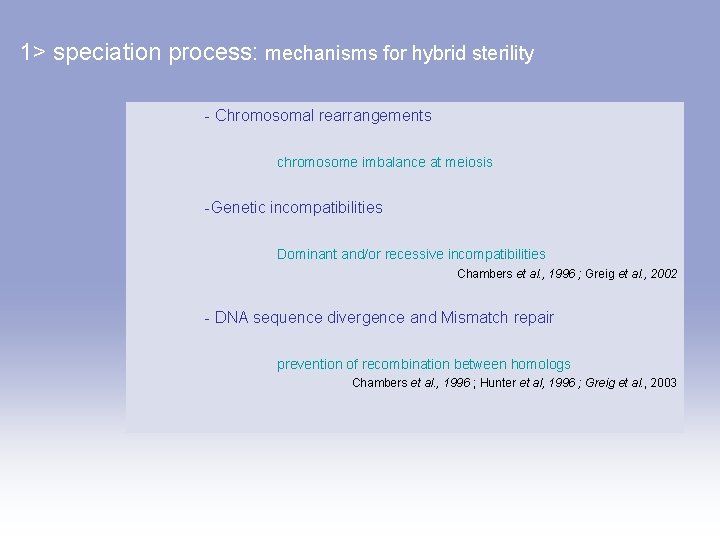
1> speciation process: mechanisms for hybrid sterility - Chromosomal rearrangements chromosome imbalance at meiosis -Genetic incompatibilities Dominant and/or recessive incompatibilities Chambers et al. , 1996 ; Greig et al. , 2002 - DNA sequence divergence and Mismatch repair prevention of recombination between homologs Chambers et al. , 1996 ; Hunter et al, 1996 ; Greig et al. , 2003

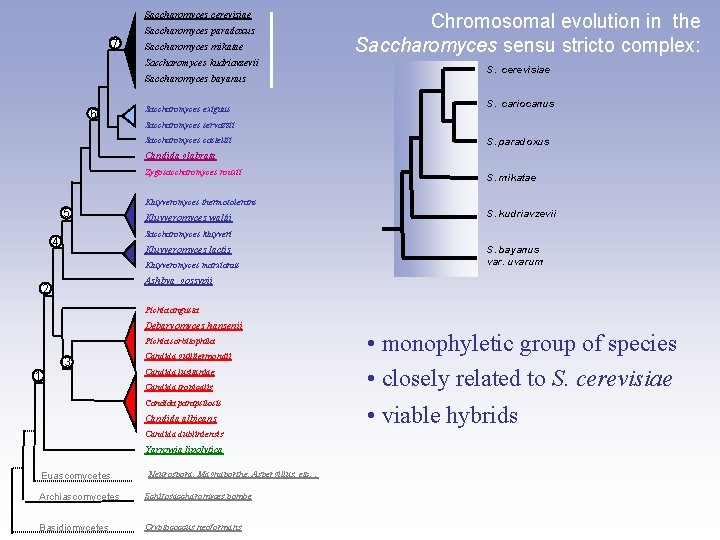
Saccharomyces cerevisiae Saccharomyces paradoxus 7 Saccharomyces mikatae Saccharomyces kudriavzevii Saccharomyces bayanus 6 Saccharomyces exiguus Chromosomal evolution in the Saccharomyces sensu stricto complex: S. cerevisiae S. cariocanus Saccharomyces servazzii Saccharomyces castellii S. paradoxus Candida glabrata Zygosaccharomyces rouxii 5 S. mikatae Kluyveromyces thermotolerans Kluyveromyces waltii S. kudriavzevii Saccharomyces kluyveri 4 Kluyveromyces lactis Kluyveromyces marxianus S. bayanus var. uvarum Ashbya gossypii 2 Pichia angusta Debaryomyces hansenii Pichia sorbitophila 3 1 Candida guilliermondii Candida lusitaniae Candida tropicalis Candida parapsilosis Candida albicans Candida dubliniensis Yarrowia lipolytica Euascomycetes Neurospora, Magnaporthe, Aspergillus, etc… Archiascomycetes Schizosaccharomyces pombe Basidiomycetes Cryptococcus neoformans • monophyletic group of species • closely related to S. cerevisiae • viable hybrids

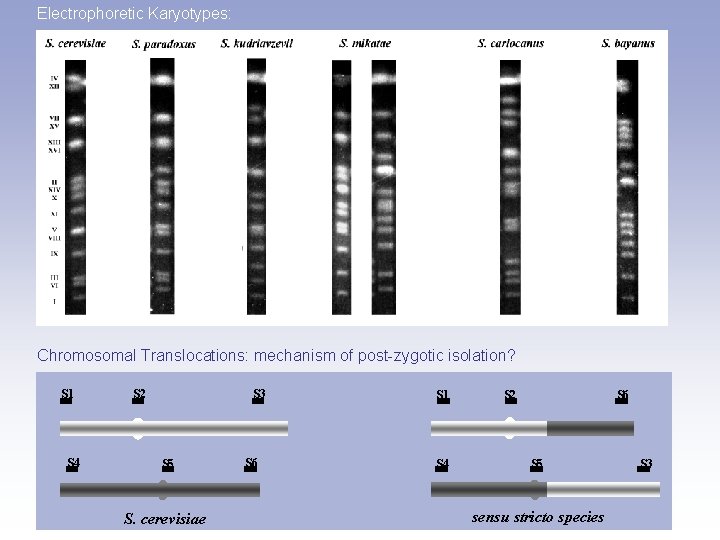
Electrophoretic Karyotypes: Chromosomal Translocations: mechanism of post-zygotic isolation? S 1 S 4 S 2 S 3 S 5 S. cerevisiae S 6 S 1 S 4 S 2 S 6 S 5 sensu stricto species S 3

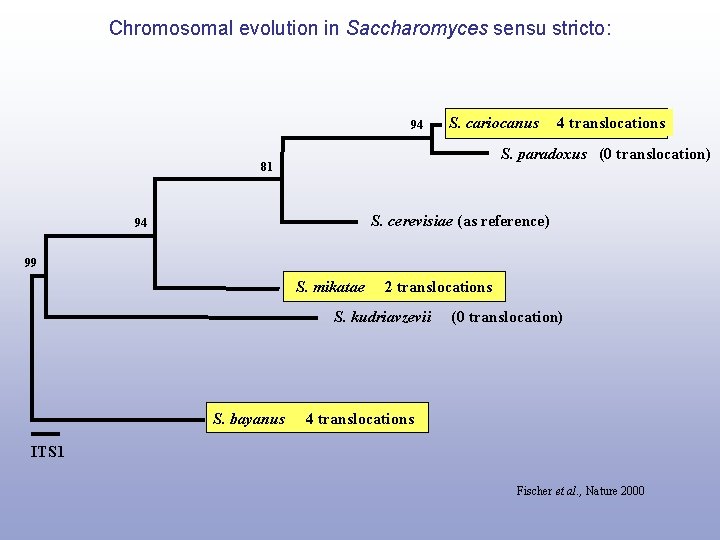
Chromosomal evolution in Saccharomyces sensu stricto: 94 S. cariocanus 4 translocations S. paradoxus (0 translocation) 81 S. cerevisiae (as reference) 94 99 S. mikatae 2 translocations S. kudriavzevii S. bayanus (0 translocation) 4 translocations ITS 1 Fischer et al. , Nature 2000

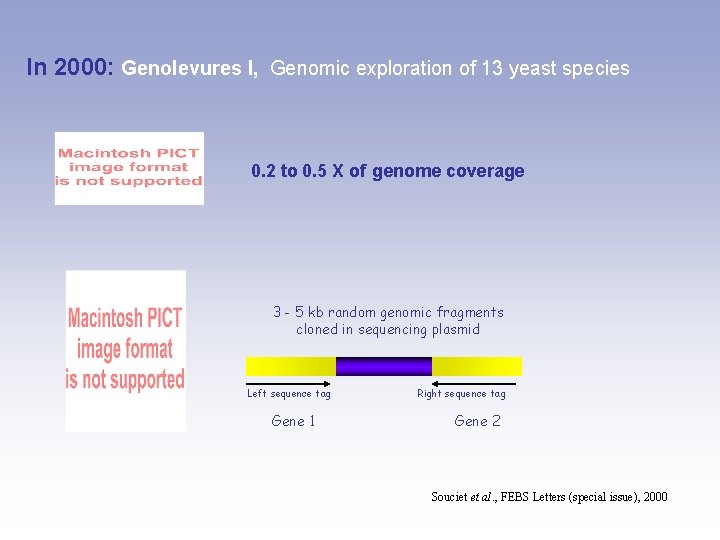
In 2000: Genolevures I, Genomic exploration of 13 yeast species 0. 2 to 0. 5 X of genome coverage 3 - 5 kb random genomic fragments cloned in sequencing plasmid Left sequence tag Gene 1 Right sequence tag Gene 2 Souciet et al. , FEBS Letters (special issue), 2000

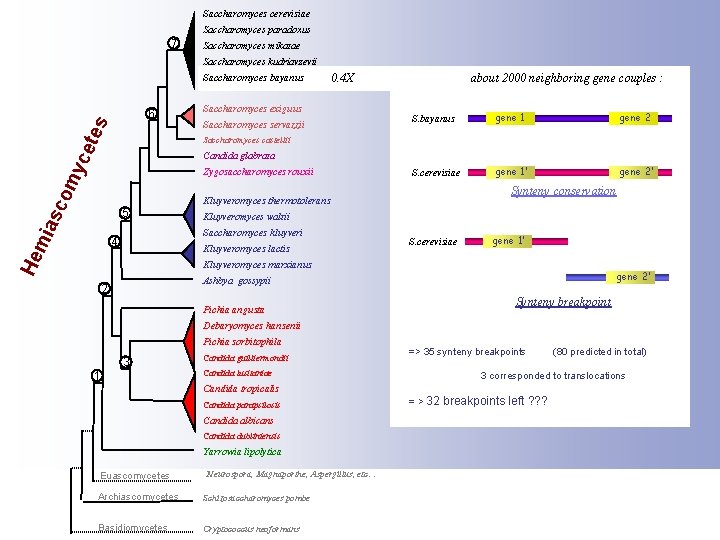
Saccharomyces cerevisiae Saccharomyces paradoxus 7 Saccharomyces mikatae Saccharomyces kudriavzevii Saccharomyces bayanus tes 6 0. 4 X Saccharomyces exiguus Saccharomyces servazzii about 2000 neighboring gene couples : S. bayanus gene 1 gene 2 S. cerevisiae gene 1’ gene 2’ yce Saccharomyces castellii Candida glabrata asc om Zygosaccharomyces rouxii mi 5 Kluyveromyces waltii Saccharomyces kluyveri 4 Kluyveromyces lactis He Synteny conservation Kluyveromyces thermotolerans S. cerevisiae gene 1’ Kluyveromyces marxianus gene 2’ Ashbya gossypii 2 Synteny breakpoint Pichia angusta Debaryomyces hansenii Pichia sorbitophila 3 1 Candida guilliermondii => 35 synteny breakpoints Candida lusitaniae 3 corresponded to translocations Candida tropicalis Candida parapsilosis Candida albicans Candida dubliniensis Yarrowia lipolytica Euascomycetes Neurospora, Magnaporthe, Aspergillus, etc… Archiascomycetes Schizosaccharomyces pombe Basidiomycetes Cryptococcus neoformans (80 predicted in total) = > 32 breakpoints left ? ? ?

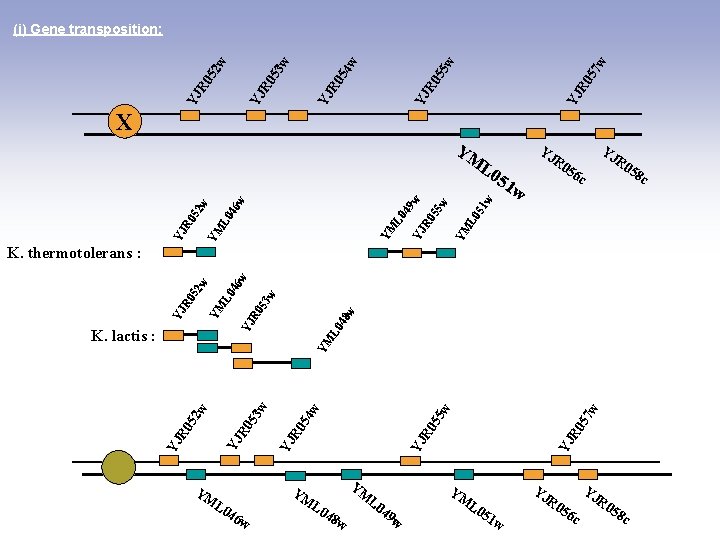
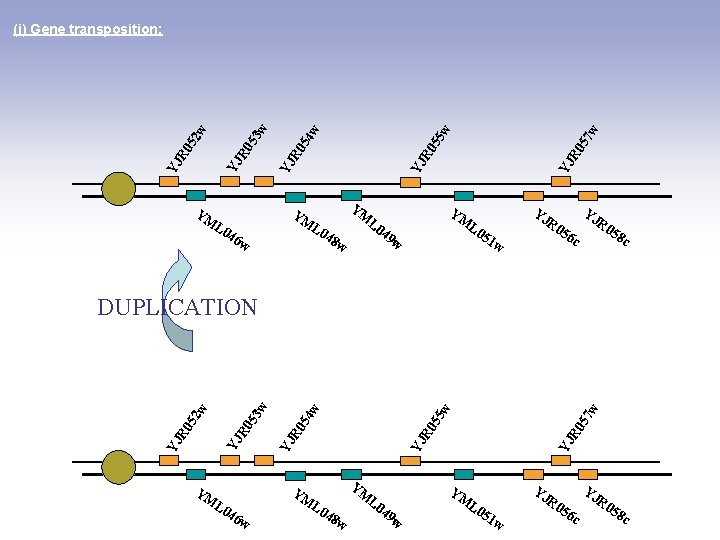
YM L 0 46 w YM L 0 YM 48 L 0 w 49 w w 57 R 0 YJ w 55 R 0 YJ w 54 R 0 YJ w 53 48 w L 0 YM 3 w 46 w L 0 YM 2 w R 0 5 YJ K. lactis : R 0 YJ w 52 R 0 YJ w 49 w YM L 0 51 55 w YJ R 0 YM L 0 46 w 52 w YJ R 0 YJ w w 57 55 R 0 YJ w w w 53 54 R 0 YJ 52 R 0 YJ (i) Gene transposition: X YM L 0 51 w YJ w R 0 56 c YJ YJ R 0 58 c K. thermotolerans : R 0 58 c

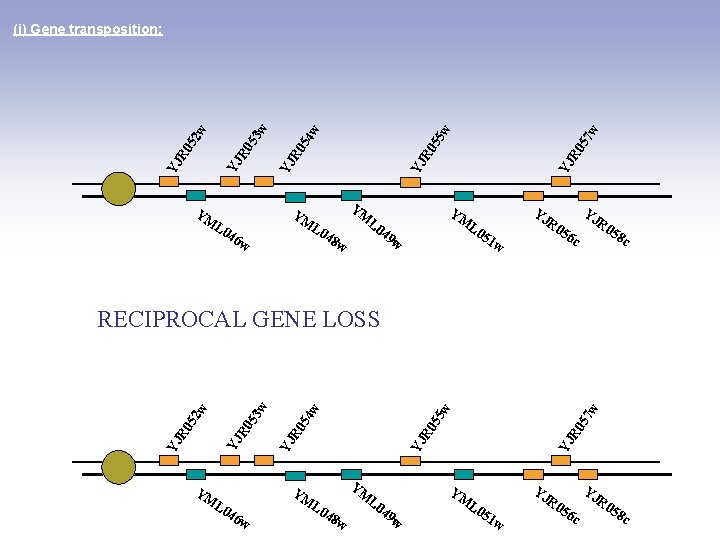
YM YM YM L 0 46 w L 0 w 7 w YJ YM L 0 49 48 YJ R 0 5 5 w YJ R 0 5 4 w YJ R 0 5 2 w 3 w (i) Gene transposition: R 0 56 c 51 w w YJ R 0 58 c YM YM YM L 0 46 w L 0 48 L 0 w w YJ R 0 57 w YJ R 0 55 w 54 R 0 YJ YJ YJ R 0 52 53 w w DUPLICATION YJ 49 YM w L 0 51 w R 0 56 c YJ R 0 58 c

YM YM YM L 0 46 w L 0 7 w L 0 49 R 0 56 c 51 w w YJ YM L 0 48 YJ R 0 5 5 w YJ R 0 5 4 w YJ R 0 5 2 w 3 w (i) Gene transposition: w YJ R 0 58 c YM YM YM L 0 46 w L 0 48 L 0 w w YJ R 0 57 w YJ R 0 55 w 54 R 0 YJ YJ YJ R 0 52 53 w w RECIPROCAL GENE LOSS YJ 49 YM w L 0 51 w R 0 56 c YJ R 0 58 c

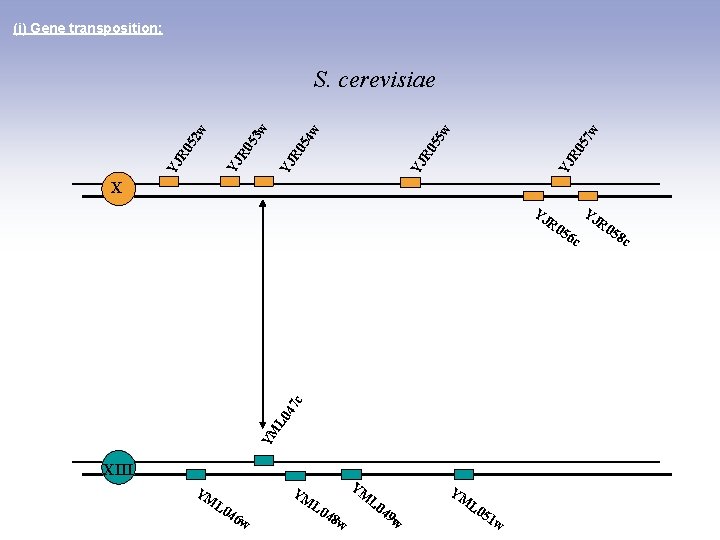
(i) Gene transposition: 7 w YJ R 0 5 5 w YJ R 0 5 4 w YJ R 0 5 2 w 3 w S. cerevisiae X YJ YM L 0 47 c R 0 56 c XIII YM YM L 0 46 YM w L 0 48 L 0 w 49 YM w L 0 51 w YJ R 0 58 c

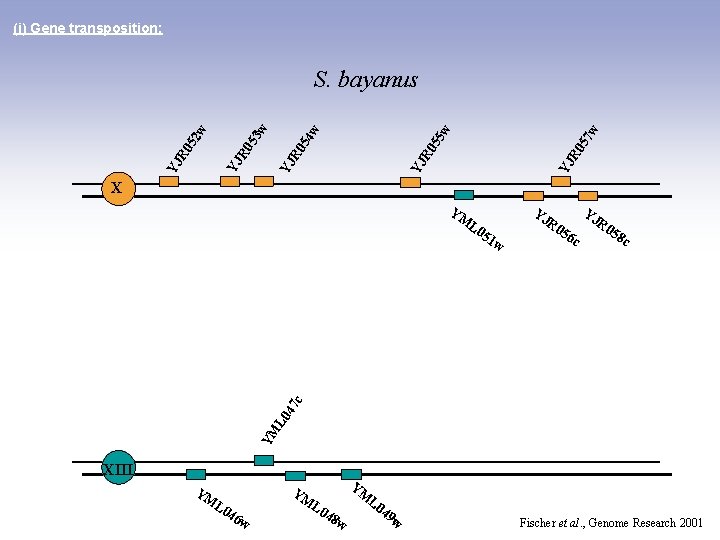
(i) Gene transposition: 7 w YJ R 0 5 5 w YJ R 0 5 4 w YJ R 0 5 2 w 3 w S. bayanus X YM L 0 YJ 51 YJ R 0 58 c YM L 0 47 c w R 0 56 c XIII YM YM L 0 46 YM w L 0 48 w 49 w Fischer et al. , Genome Research 2001

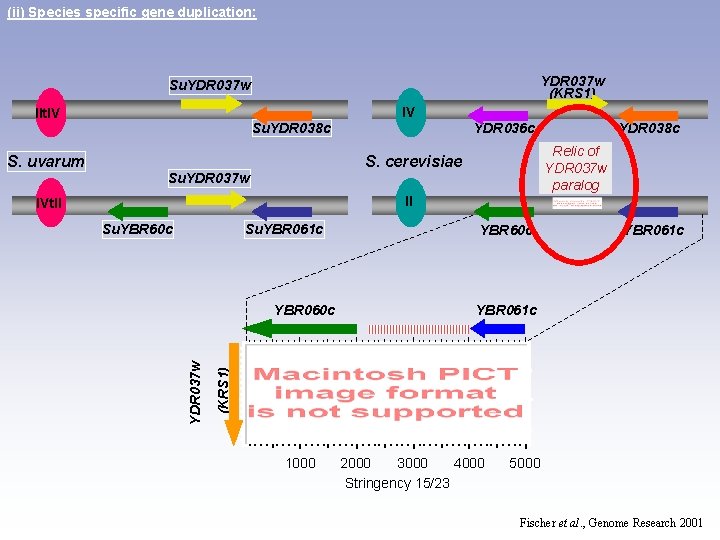
(ii) Species specific gene duplication: YDR 037 w (KRS 1) Su. YDR 037 w IV IIt. IV Su. YDR 038 c S. uvarum YDR 036 c YDR 038 c Relic of YDR 037 w paralog S. cerevisiae Su. YDR 037 w II IVt. II Su. YBR 60 c Su. YBR 061 c (KRS 1) YDR 037 w YBR 060 c YBR 60 c 1000 2000 3000 4000 Stringency 15/23 5000 Fischer et al. , Genome Research 2001

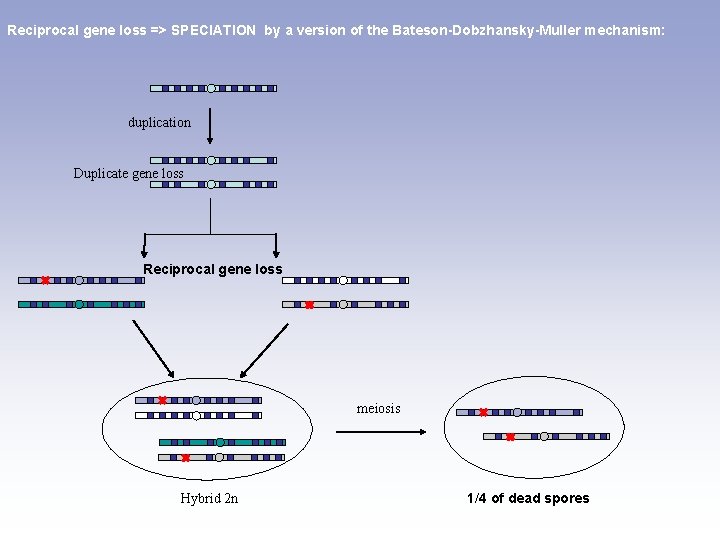
Reciprocal gene loss => SPECIATION by a version of the Bateson-Dobzhansky-Muller mechanism: duplication Duplicate gene loss Reciprocal gene loss meiosis Hybrid 2 n 1/4 of dead spores

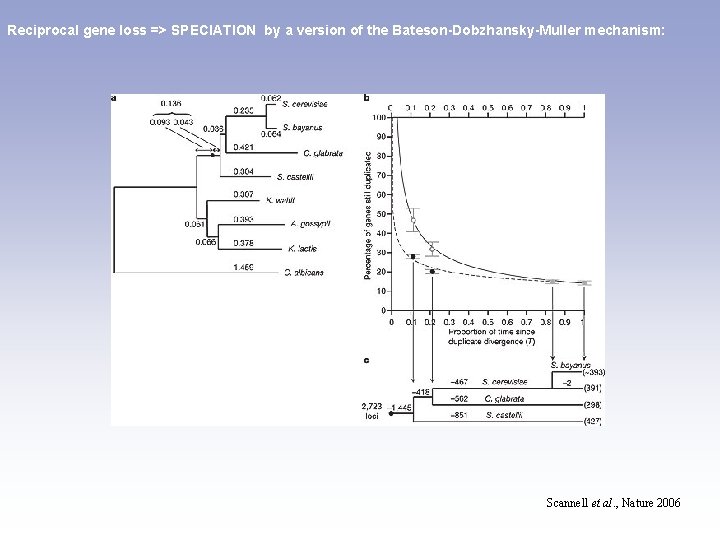
Reciprocal gene loss => SPECIATION by a version of the Bateson-Dobzhansky-Muller mechanism: Scannell et al. , Nature 2006

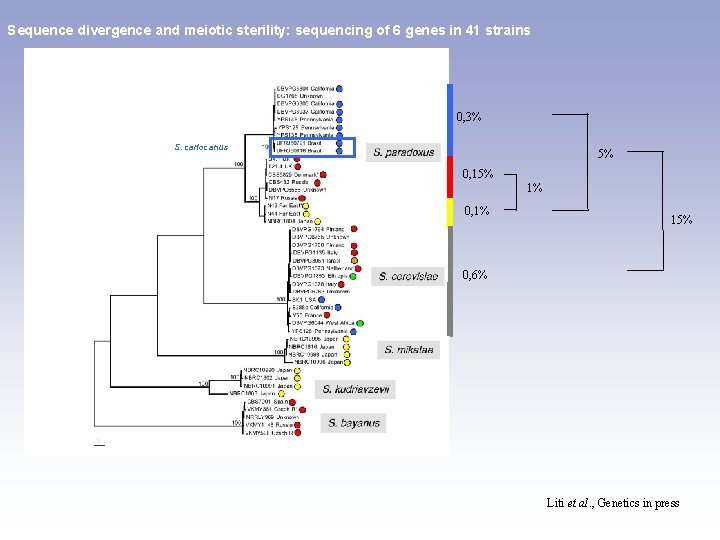
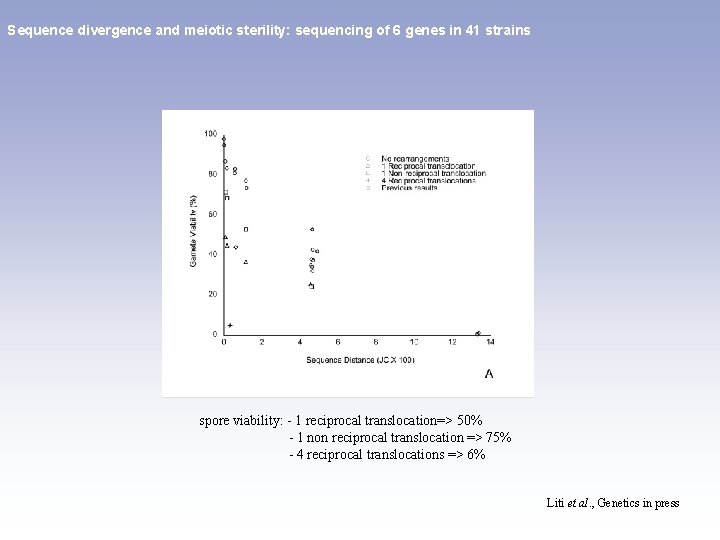
Sequence divergence and meiotic sterility: sequencing of 6 genes in 41 strains 0, 3% S. cariocanus 5% 0, 15% 1% 0, 1% 15% 0, 6% Liti et al. , Genetics in press

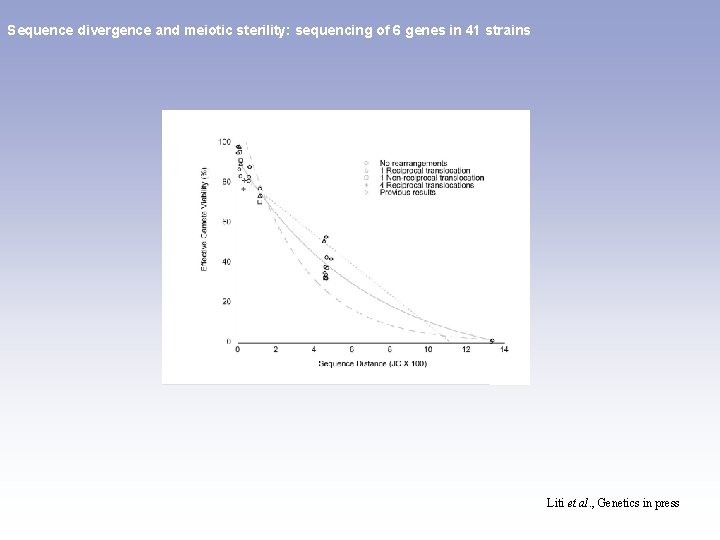
Sequence divergence and meiotic sterility: sequencing of 6 genes in 41 strains spore viability: - 1 reciprocal translocation=> 50% - 1 non reciprocal translocation => 75% - 4 reciprocal translocations => 6% Liti et al. , Genetics in press

Sequence divergence and meiotic sterility: sequencing of 6 genes in 41 strains Liti et al. , Genetics in press

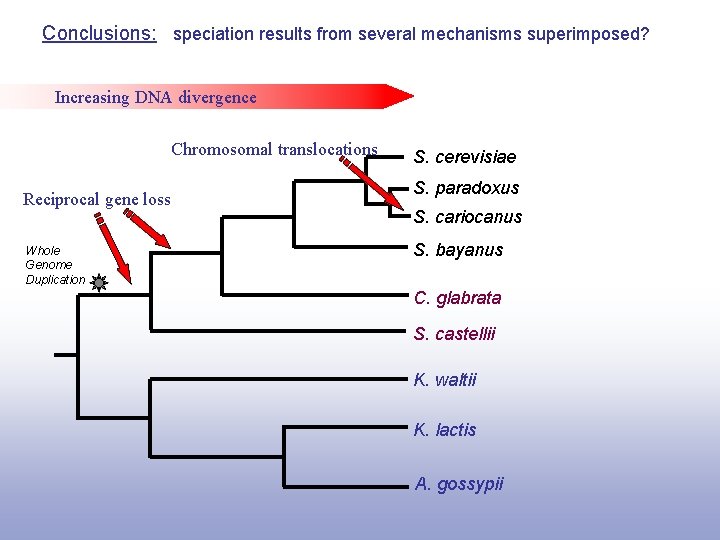
Conclusions: speciation results from several mechanisms superimposed? Increasing DNA divergence Chromosomal translocations Reciprocal gene loss Whole Genome Duplication S. cerevisiae S. paradoxus S. cariocanus S. bayanus C. glabrata S. castellii K. waltii K. lactis A. gossypii

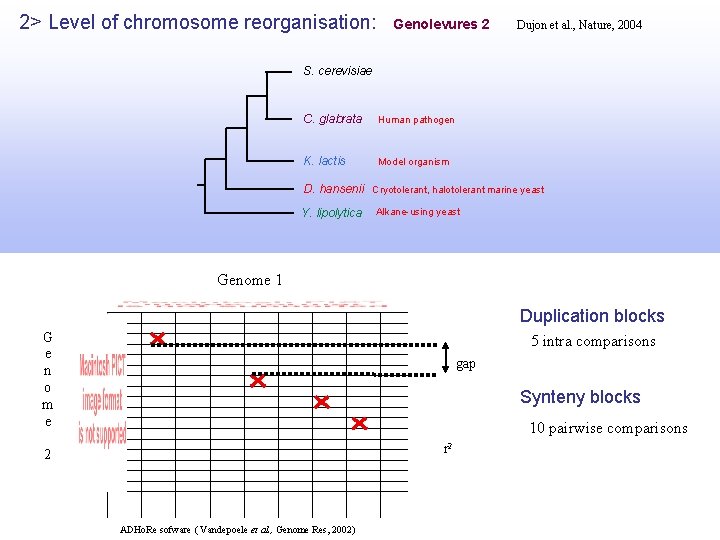
2> Level of chromosome reorganisation: Genolevures 2 Dujon et al. , Nature, 2004 S. cerevisiae C. glabrata Human pathogen K. lactis Model organism D. hansenii Cryotolerant, halotolerant marine yeast Y. lipolytica Alkane-using yeast Genome 1 Duplication blocks G e n o m e 5 intra comparisons gap Synteny blocks 10 pairwise comparisons r 2 2 ADHo. Re sofware ( Vandepoele et al. , Genome Res, 2002)

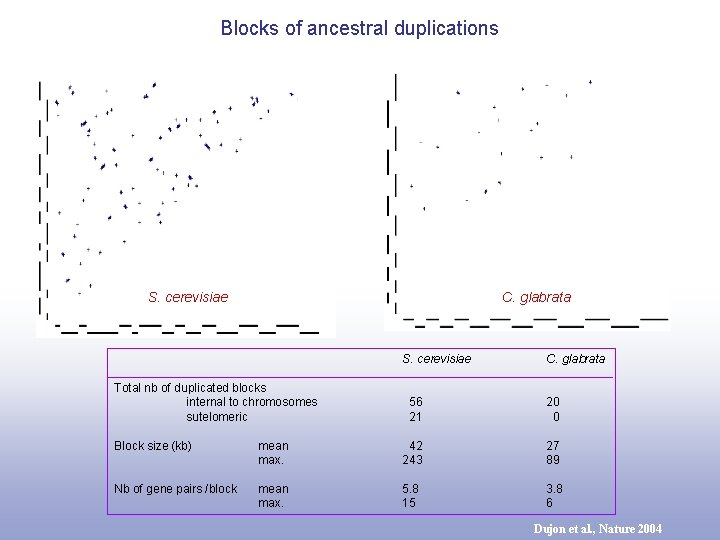
Blocks of ancestral duplications S. cerevisiae C. glabrata S. cerevisiae Total nb of duplicated blocks internal to chromosomes sutelomeric C. glabrata 56 21 20 0 Block size (kb) mean max. 42 243 27 89 Nb of gene pairs /block mean max. 5. 8 15 3. 8 6 Dujon et al. , Nature 2004

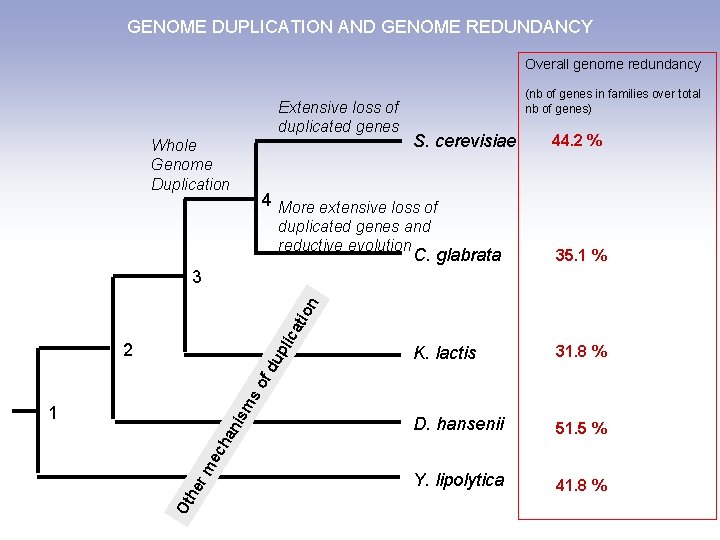
GENOME DUPLICATION AND GENOME REDUNDANCY Overall genome redundancy Extensive loss of duplicated genes Whole Genome Duplication (nb of genes in families over total nb of genes) S. cerevisiae 44. 2 % 4 More extensive loss of duplicated genes and reductive evolution C. glabrata 35. 1 % K. lactis 31. 8 % D. hansenii 51. 5 % Y. lipolytica 41. 8 % ca tio n 3 ms of du pli 2 Ot he rm ec ha nis 1

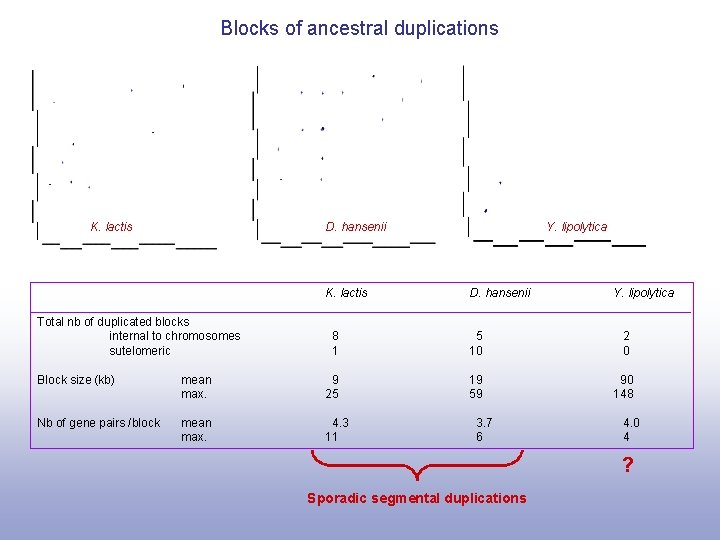
Blocks of ancestral duplications K. lactis D. hansenii K. lactis Total nb of duplicated blocks internal to chromosomes sutelomeric Y. lipolytica D. hansenii Y. lipolytica 8 1 5 10 2 0 19 59 90 148 Block size (kb) mean max. 9 25 Nb of gene pairs /block mean max. 4. 3 11 3. 7 6 4. 0 4 ? Sporadic segmental duplications

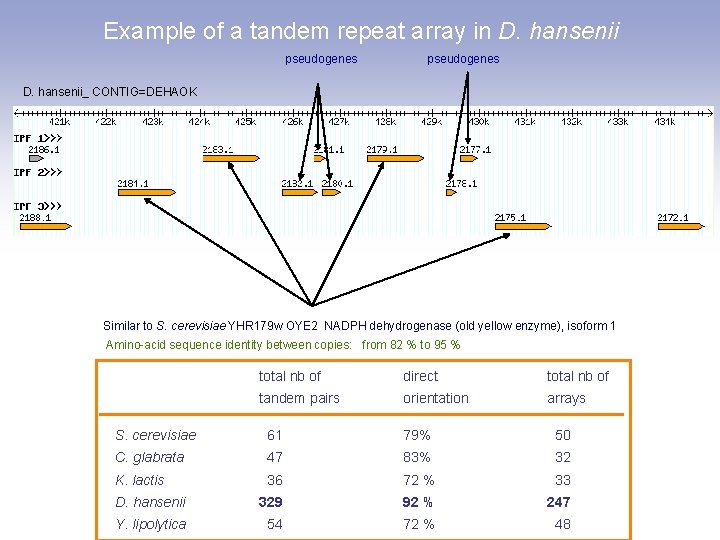
Example of a tandem repeat array in D. hansenii pseudogenes D. hansenii_ CONTIG=DEHAOK Similar to S. cerevisiae YHR 179 w OYE 2 NADPH dehydrogenase (old yellow enzyme), isoform 1 Amino-acid sequence identity between copies: from 82 % to 95 % total nb of direct total nb of tandem pairs orientation arrays S. cerevisiae 61 79% 50 C. glabrata 47 83% 32 K. lactis 36 72 % 33 D. hansenii 329 92 % 247 Y. lipolytica 54 72 % 48

GENOME DUPLICATION AND GENOME REDUNDANCY Extensive loss of duplicated genes Whole Genome Duplication S. cerevisiae Segmental duplication 4 More extensive loss of duplicated genes and reductive evolution C. glabrata 3 2 1 Segmental duplication K. lactis Tandem repeat formation Segmental duplication D. hansenii Y. lipolytica

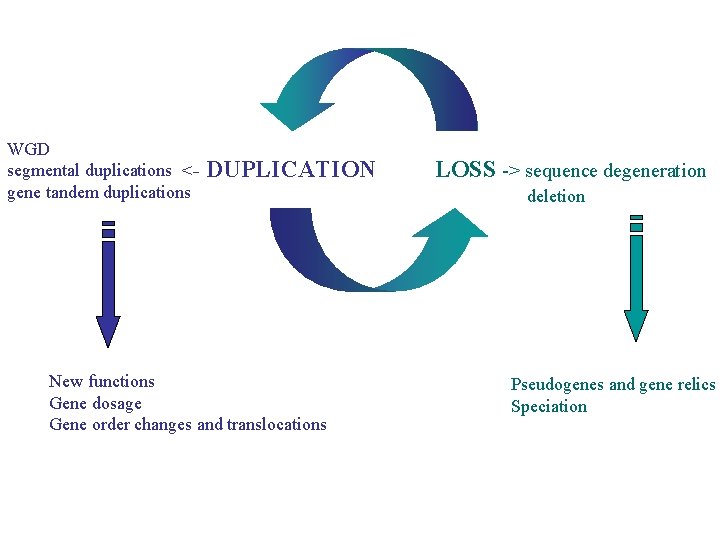
WGD segmental duplications <gene tandem duplications DUPLICATION New functions Gene dosage Gene order changes and translocations LOSS -> sequence degeneration deletion Pseudogenes and gene relics Speciation

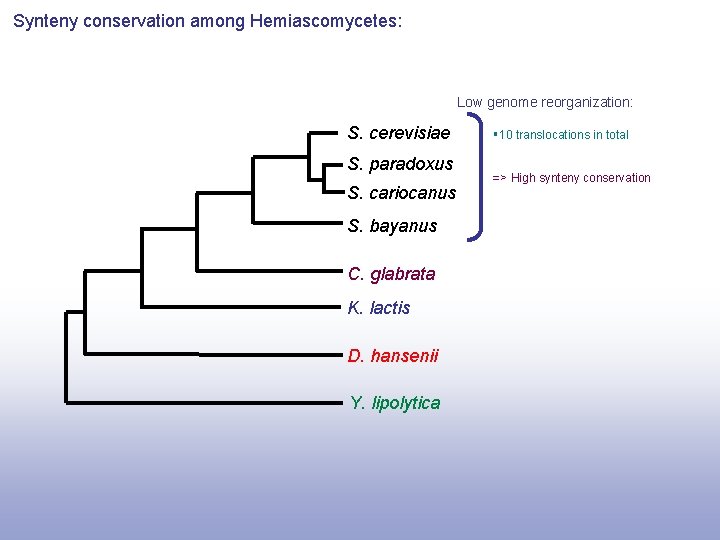
Synteny conservation among Hemiascomycetes: Low genome reorganization: S. cerevisiae S. paradoxus S. cariocanus S. bayanus C. glabrata K. lactis D. hansenii Y. lipolytica § 10 translocations in total => High synteny conservation

S. cerevisiae C. glabrata K. lactis D. hansenii Y. lipolytica C. glabrata K. lactis D. hansenii

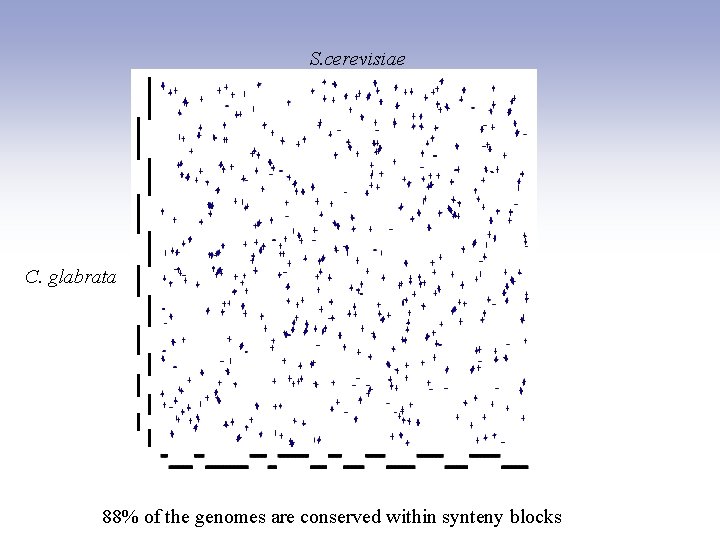
S. cerevisiae C. glabrata 88% of the genomes are conserved within synteny blocks


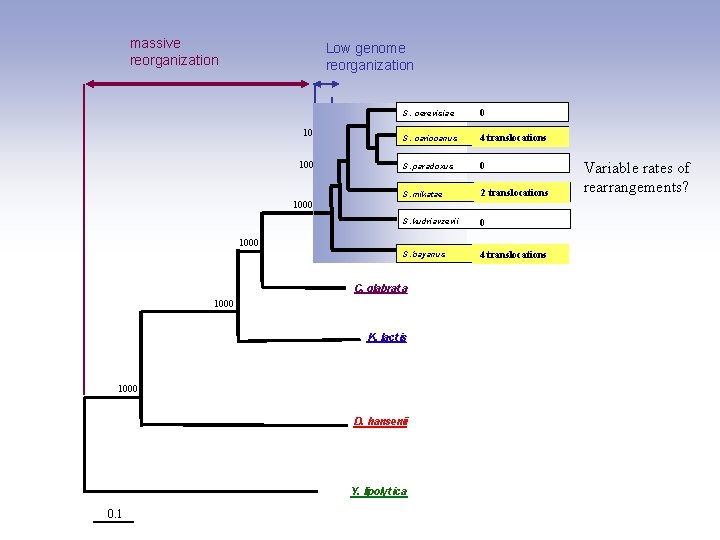
massive reorganization Low genome reorganization S. S. cerevisiae 1000 cerevisiae S. cariocanus S. paradoxus S. mikatae 1000 0 reference 4 translocations 0 2 translocations S. mikatae S. kudriavzevii 0 1000 S. bayanus C. glabrat a 1000 K. lact is 1000 D. hansenii Y. lipolyt ica 0. 1 4 translocations Variable rates of rearrangements?

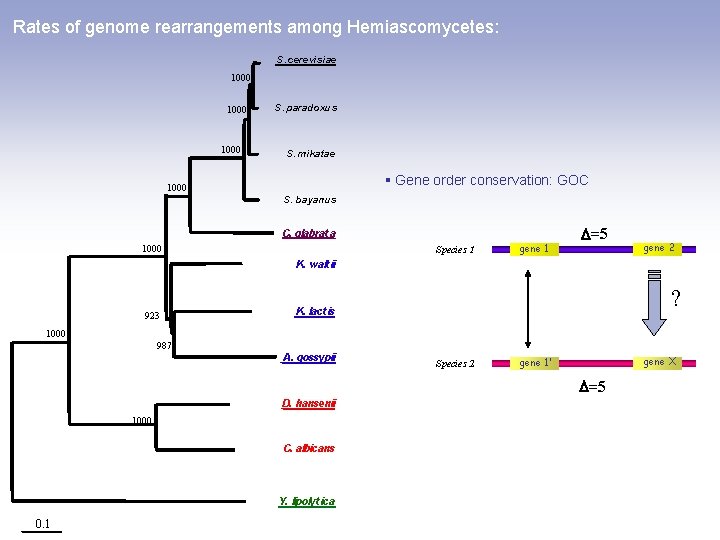
Rates of genome rearrangements among Hemiascomycetes: S. cerevisiae 1000 S. paradoxus S. mikatae § Gene order conservation: GOC 1000 S. bayanus C. glabrat a 1000 Species 1 gene 1 =5 gene 2 K. walt ii 923 ? K. lact is 1000 987 A. gossypii Species 2 gene X gene 1’ =5 D. hansenii 1000 C. albicans Y. lipolyt ica 0. 1

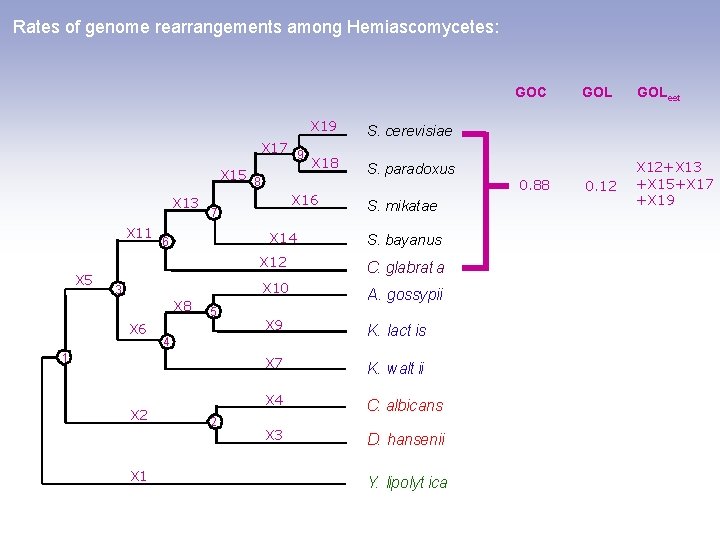
Rates of genome rearrangements among Hemiascomycetes: GOC X 17 9 X 15 8 X 13 X 11 X 5 X 14 3 X 8 X 6 5 X 2 X 18 S. paradoxus S. mikatae S. bayanus X 12 C. glabrat a X 10 A. gossypii X 9 K. lact is X 7 K. walt ii X 4 C. albicans X 3 D. hansenii 4 1 S. cerevisiae X 16 7 6 X 19 Y. lipolyt ica 0. 88 GOLest 0. 12 X 12+X 13 +X 15+X 17 +X 19

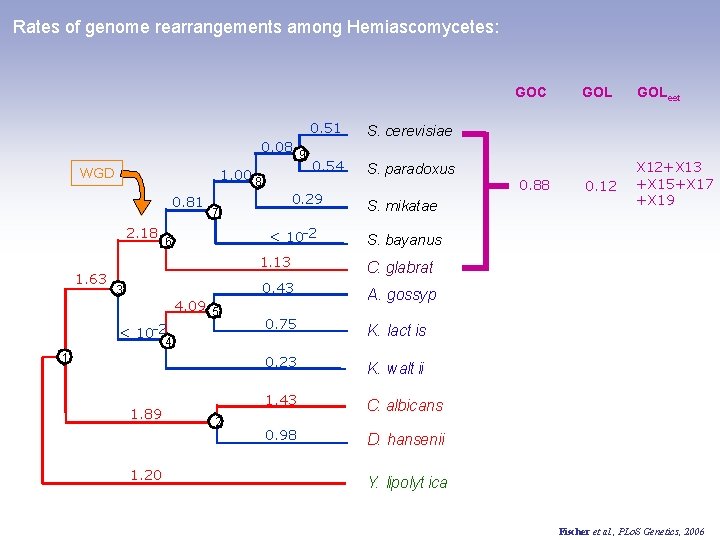
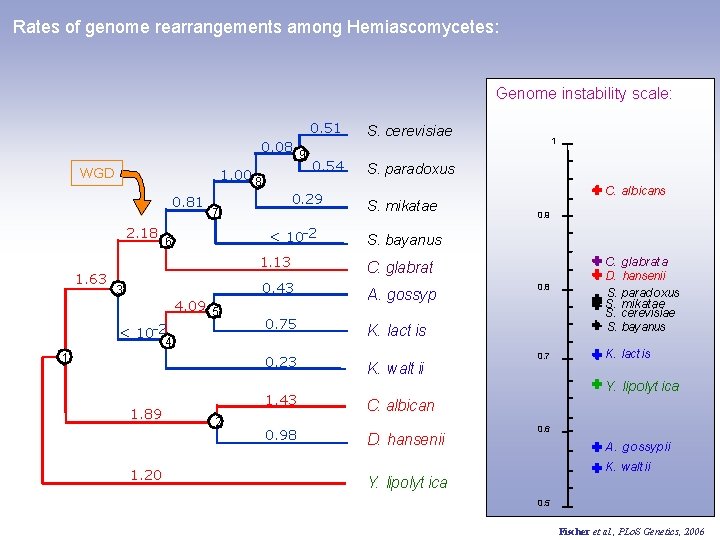
Rates of genome rearrangements among Hemiascomycetes: GOC 0. 08 9 WGD 1. 00 8 0. 81 2. 18 1. 63 0. 54 S. paradoxus < 10 -2 6 4. 09 5 < 10 -2 1 2 S. mikatae 0. 88 GOLest 0. 12 X 12+X 13 +X 15+X 17 +X 19 S. bayanus 1. 13 C. glabrat 0. 43 A. gossyp 0. 75 K. lact is 0. 23 K. walt ii 1. 43 C. albicans 0. 98 D. hansenii 4 1. 20 S. cerevisiae 0. 29 7 3 1. 89 0. 51 GOL Y. lipolyt ica Fischer et al. , PLo. S Genetics, 2006

Rates of genome rearrangements among Hemiascomycetes: Genome instability scale: 0. 08 9 WGD 1. 00 8 0. 81 2. 18 1. 63 0. 54 S. paradoxus < 10 -2 6 4. 09 5 < 10 -2 1 0. 43 A. gossyp 0. 75 K. lact is 1. 43 0. 98 1 C. albicans 0. 9 S. bayanus C. glabrat 0. 23 2 S. mikatae 1. 13 4 1. 20 S. cerevisiae 0. 29 7 3 1. 89 0. 51 K. walt ii 0. 8 0. 7 C. glabrat a D. hansenii S. paradoxus S. mikat ae S. cerevisiae S. bayanus K. lact is Y. lipolyt ica C. albican D. hansenii 0. 6 A. gossypii K. walt ii Y. lipolyt ica 0. 5 Fischer et al. , PLo. S Genetics, 2006

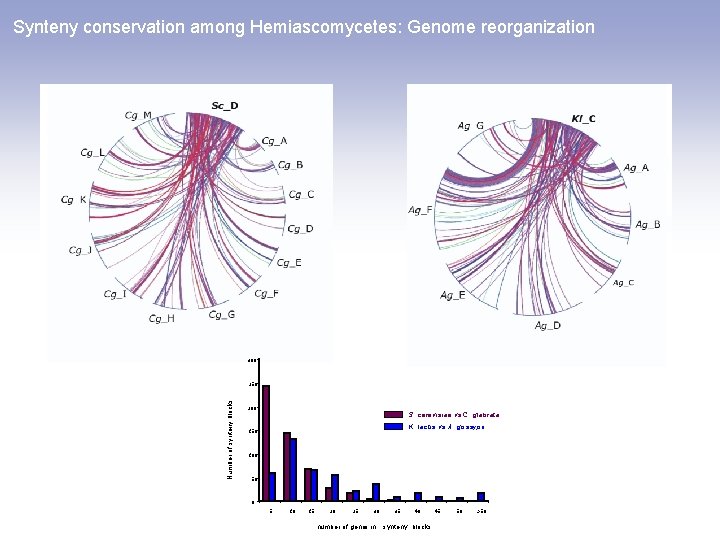
Synteny conservation among Hemiascomycetes: Genome reorganization 300 Number of synteny blocks 250 200 S. cerevisiae vs C. glabrata K. lactis vs A. gossypii 150 100 50 0 5 10 15 20 25 30 35 40 number of genes in synteny blocks 45 50 >50

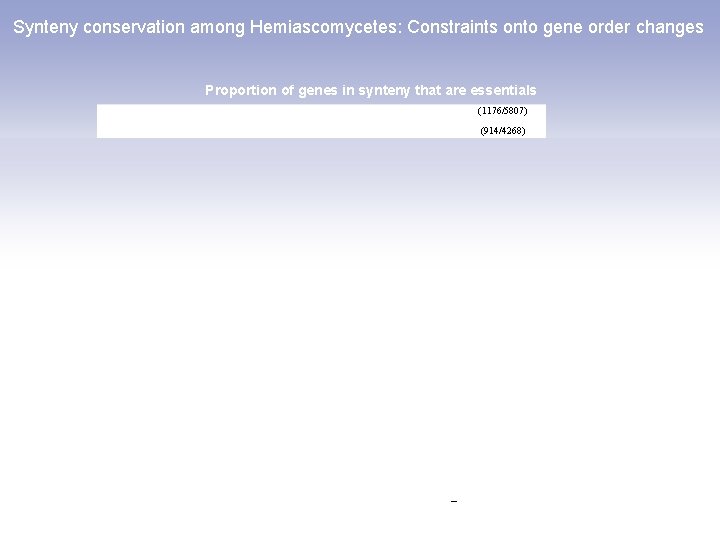
Synteny conservation among Hemiascomycetes: Constraints onto gene order changes Proportion of genes in synteny that are essentials (1176/5807) (914/4268) (886/3814) (101/340) (58/157) (625/2481) (40/113) (10/23)

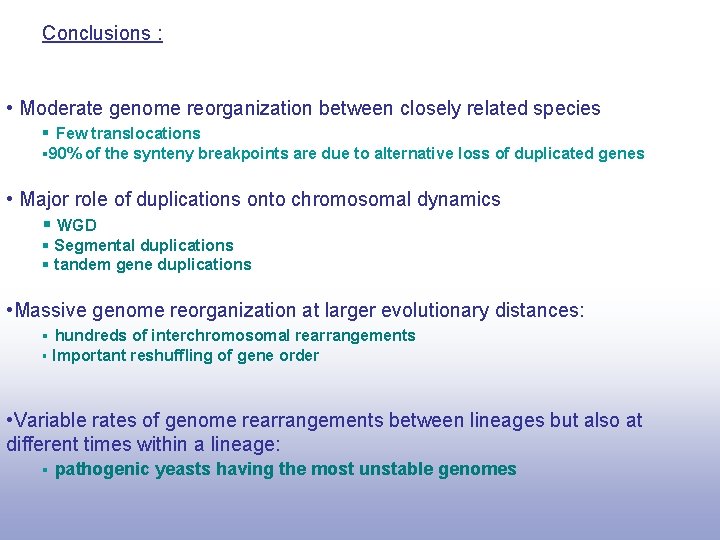
Conclusions : • Moderate genome reorganization between closely related species § Few translocations § 90% of the synteny breakpoints are due to alternative loss of duplicated genes • Major role of duplications onto chromosomal dynamics § WGD § Segmental duplications § tandem gene duplications • Massive genome reorganization at larger evolutionary distances: § hundreds of interchromosomal rearrangements § Important reshuffling of gene order • Variable rates of genome rearrangements between lineages but also at different times within a lineage: § pathogenic yeasts having the most unstable genomes

- Unité de Génétique Moléculaire des Levures Romain Koszul, Celia Payen, Ingrid Lafontaine, Bernard Dujon - The Génolevures Sequencing Consortium GDR 2354 CNRS Genoscope Plateforme séquençage, Genopole Institut Pasteur Cécile Neuvéglise Pascal Durrens Jean-Luc Souciet - Unité de Genetique des Génomes Bactériens Eduardo Rocha - Unité Génomique des Microorganismes Pathogènes Massimo Vergassola - Laboratoire de Biologie Moléculaire de la Cellule (ENS Lyon) Frédéric Brunet - Genome Stability Group (Nottinhgam, UK) Edward J. Louis
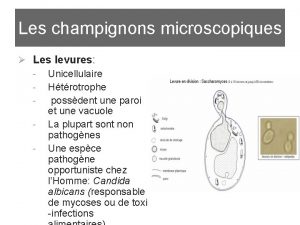
 Observation microscopique de levure et moisissure
Observation microscopique de levure et moisissure Wifi-2me
Wifi-2me Levures
Levures 2me mains
2me mains If garden gnomes regain popularity, what will happen?
If garden gnomes regain popularity, what will happen? Gnomes united case study answers
Gnomes united case study answers Victorian garden gnomes
Victorian garden gnomes Des des des
Des des des Evolution des vetements
Evolution des vetements Frise chronologique évolution des robots
Frise chronologique évolution des robots Evolution des tracteurs
Evolution des tracteurs Evolution des ponts
Evolution des ponts évolution des ponts
évolution des ponts Evolution des pcs
Evolution des pcs Evolution des ponts
Evolution des ponts Budget des ventes méthode des moindres carrés
Budget des ventes méthode des moindres carrés Diversification des espaces et des acteurs de la production
Diversification des espaces et des acteurs de la production Je t'offrirai des fleurs et des nappes en couleurs
Je t'offrirai des fleurs et des nappes en couleurs Il existe des personnes qui sont des lumières pour tous
Il existe des personnes qui sont des lumières pour tous Volume des liquides et des solides
Volume des liquides et des solides Robin des bois des alpes
Robin des bois des alpes Valeur de mode
Valeur de mode Mesure de niveaux pour liquides et solides
Mesure de niveaux pour liquides et solides Triple des attack
Triple des attack Des phrases et des bases affiche soviétique 1952
Des phrases et des bases affiche soviétique 1952 Cartographie des risques gestion des stocks
Cartographie des risques gestion des stocks La diffusion des idées des lumières
La diffusion des idées des lumières Star life cycle 1-10
Star life cycle 1-10 Terrain evolution
Terrain evolution Product evolution
Product evolution Whale evolution chart
Whale evolution chart Frog digestive system diagram
Frog digestive system diagram Wurcan
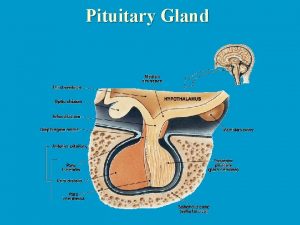
Wurcan Prolactin hormones
Prolactin hormones Chapter 7 the evolution of living things answers
Chapter 7 the evolution of living things answers Long term evolution advanced
Long term evolution advanced Homeobox genes worksheet
Homeobox genes worksheet Chapter 16 primate evolution
Chapter 16 primate evolution Bryophyta hepatophyta anthocerophyta
Bryophyta hepatophyta anthocerophyta Msg-3 maintenance program
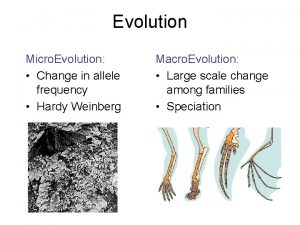
Msg-3 maintenance program Micro evolution
Micro evolution Lamarck's theory of evolution
Lamarck's theory of evolution Indirect evidence of evolution
Indirect evidence of evolution