PROTEIN SYNTHESIS How your cell makes very important





























- Slides: 54

PROTEIN SYNTHESIS

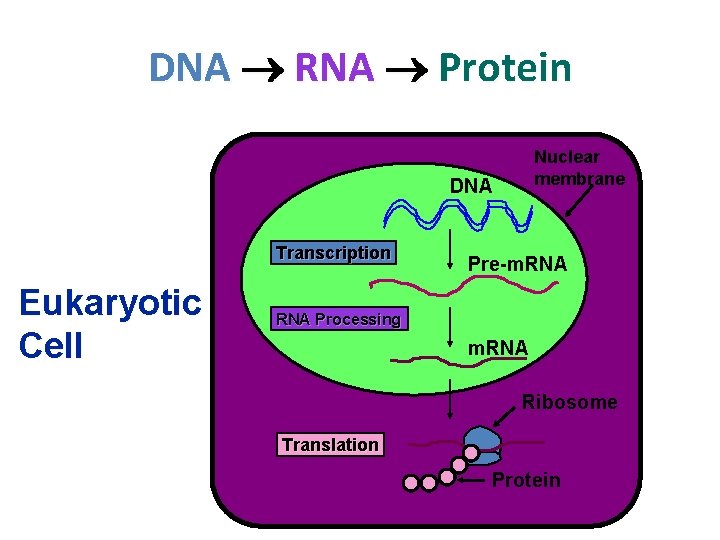
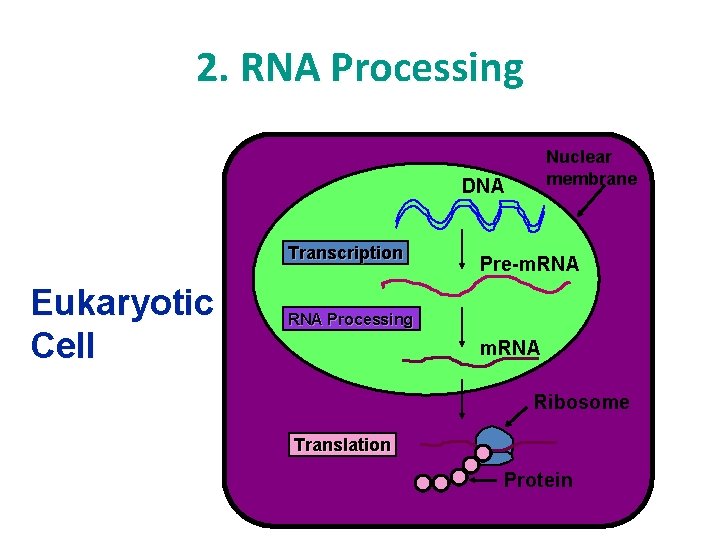
How your cell makes very important proteins • The production (synthesis) of proteins. • 3 phases: 1. Transcription 2. RNA processing 3. Translation • DNA RNA Protein

DNA RNA Protein Nuclear membrane DNA Transcription Eukaryotic Cell Pre-m. RNA Processing m. RNA Ribosome Translation Protein

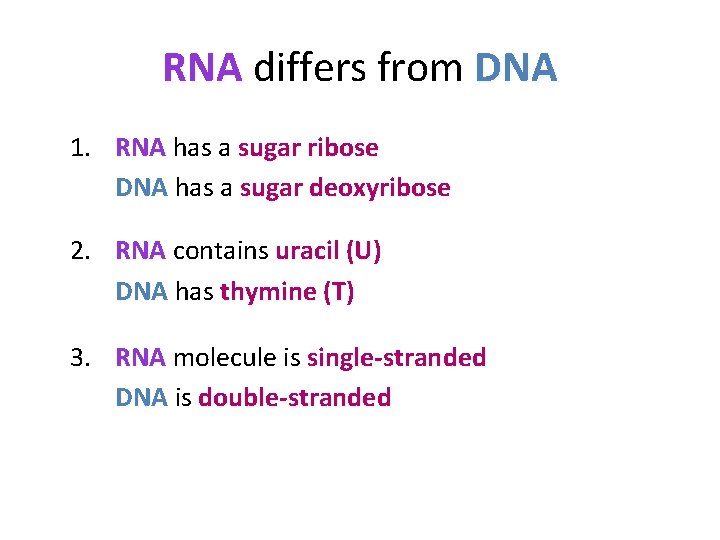
RNA differs from DNA 1. RNA has a sugar ribose DNA has a sugar deoxyribose 2. RNA contains uracil (U) DNA has thymine (T) 3. RNA molecule is single-stranded DNA is double-stranded


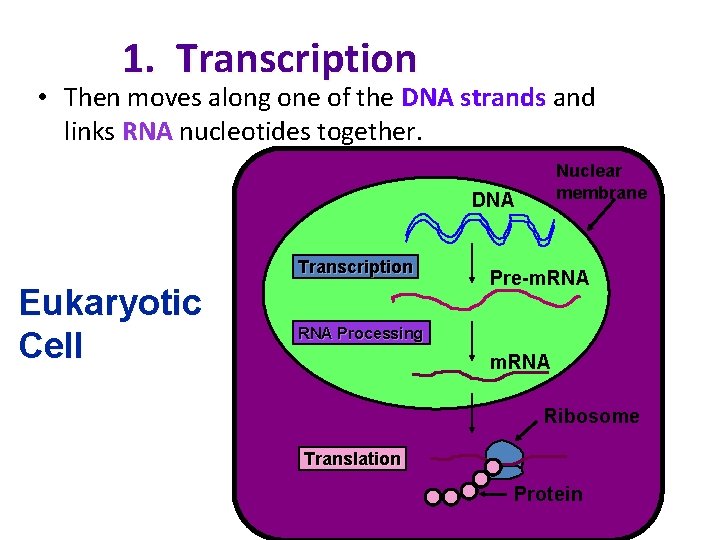
1. Transcription • Then moves along one of the DNA strands and links RNA nucleotides together. Nuclear membrane DNA Transcription Eukaryotic Cell Pre-m. RNA Processing m. RNA Ribosome Translation Protein

2. RNA Processing Nuclear membrane DNA Transcription Eukaryotic Cell Pre-m. RNA Processing m. RNA Ribosome Translation Protein

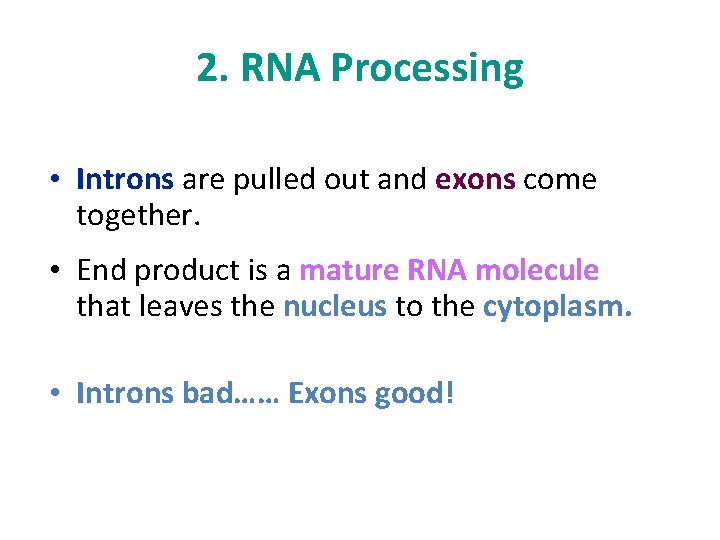
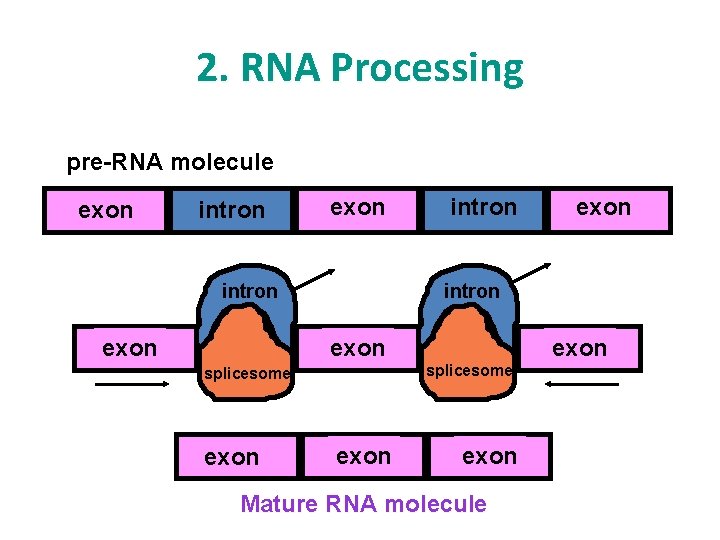
2. RNA Processing • Introns are pulled out and exons come together. • End product is a mature RNA molecule that leaves the nucleus to the cytoplasm. • Introns bad…… Exons good!

2. RNA Processing pre-RNA molecule exon intron exon splicesome exon Mature RNA molecule

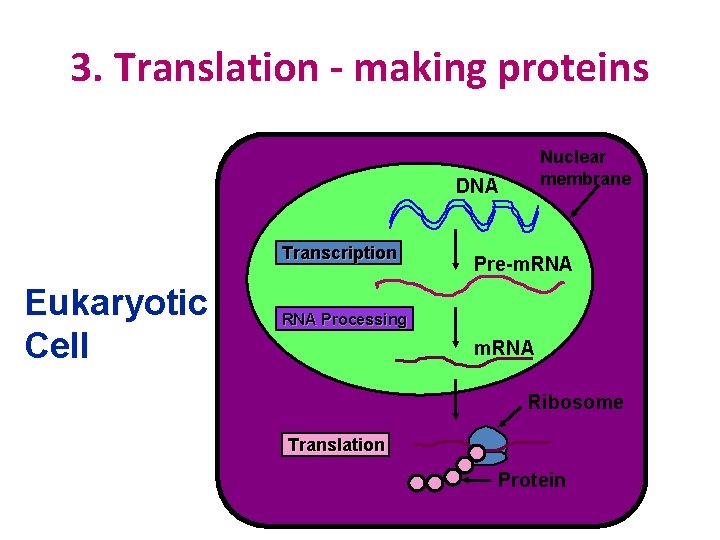
3. Translation - making proteins Nuclear membrane DNA Transcription Eukaryotic Cell Pre-m. RNA Processing m. RNA Ribosome Translation Protein















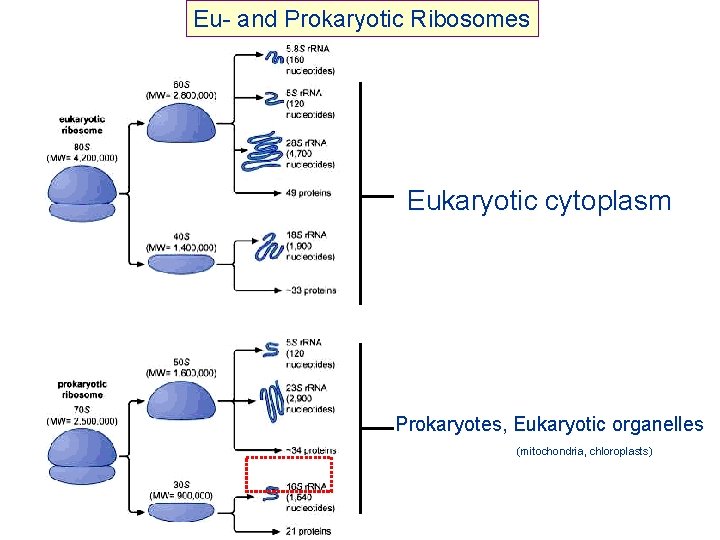
Eu- and Prokaryotic Ribosomes Eukaryotic cytoplasm Prokaryotes, Eukaryotic organelles (mitochondria, chloroplasts)

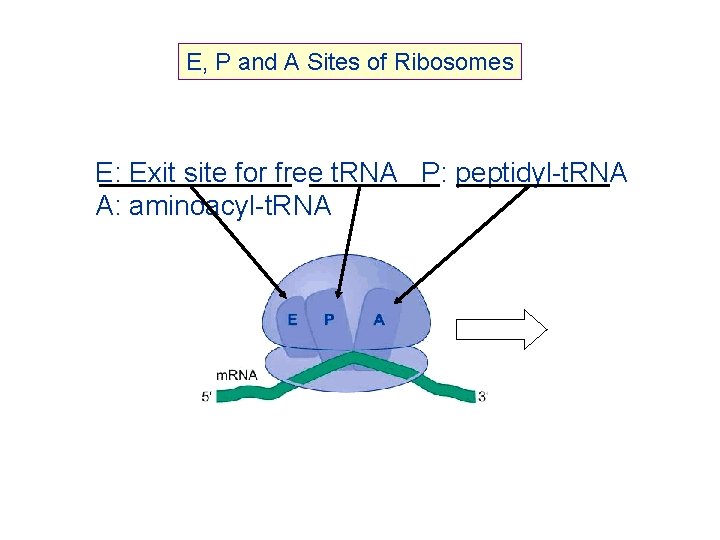
E, P and A Sites of Ribosomes E: Exit site for free t. RNA P: peptidyl-t. RNA A: aminoacyl-t. RNA

Translation • Three parts: 1. Initiation: start codon (AUG) 2. Elongation: 3. Termination: stop codon (UAG)

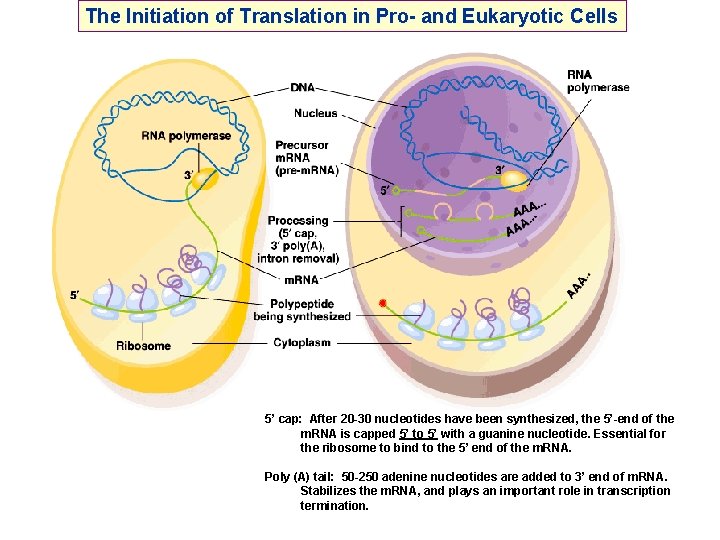
The Initiation of Translation in Pro- and Eukaryotic Cells 5’ cap: After 20 -30 nucleotides have been synthesized, the 5’-end of the m. RNA is capped 5’ to 5’ with a guanine nucleotide. Essential for the ribosome to bind to the 5’ end of the m. RNA. Poly (A) tail: 50 -250 adenine nucleotides are added to 3’ end of m. RNA. Stabilizes the m. RNA, and plays an important role in transcription termination.



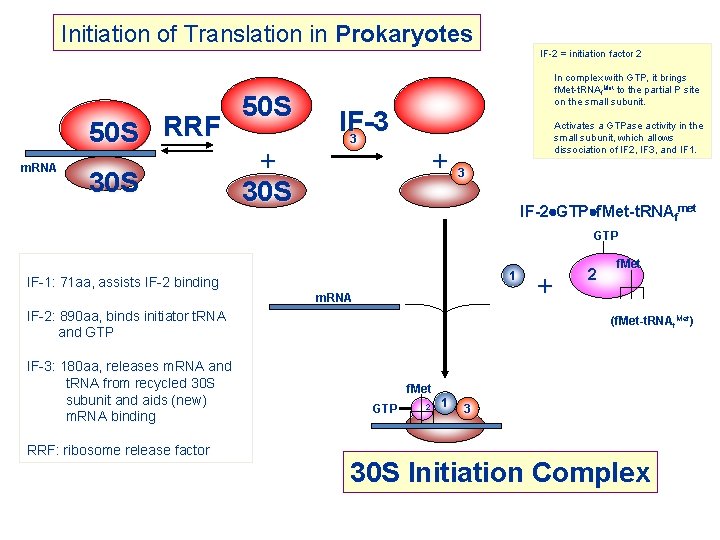
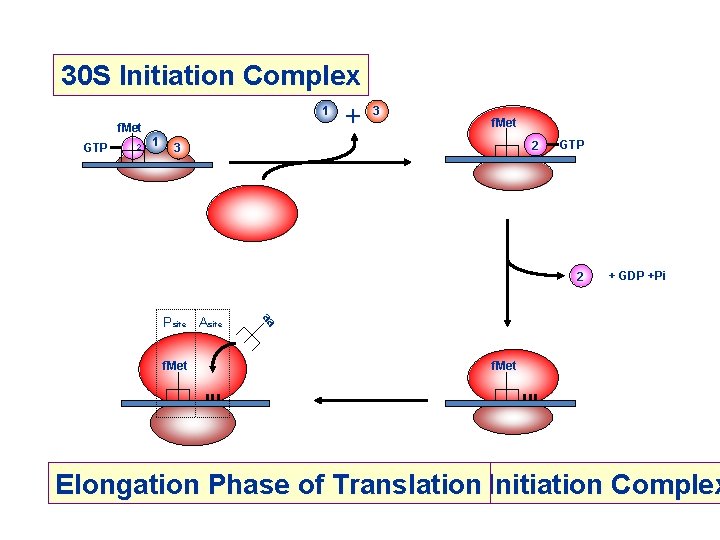
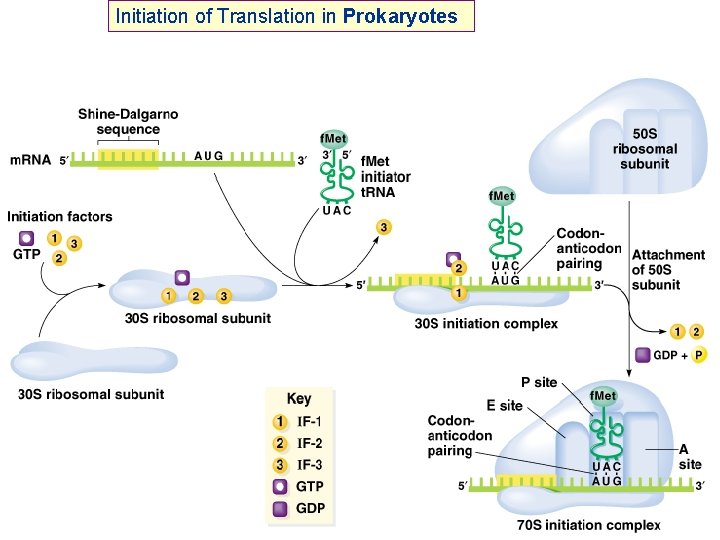
Initiation of Translation in Prokaryotes 50 S RRF m. RNA 30 S 50 S + 30 S IF-2 = initiation factor 2 In complex with GTP, it brings f. Met-t. RNAf. Met to the partial P site on the small subunit. IF-3 3 Activates a GTPase activity in the small subunit, which allows dissociation of IF 2, IF 3, and IF 1. +3 IF-2 GTP f. Met-t. RNAfmet GTP 1 IF-1: 71 aa, assists IF-2 binding m. RNA IF-2: 890 aa, binds initiator t. RNA and GTP IF-3: 180 aa, releases m. RNA and t. RNA from recycled 30 S subunit and aids (new) m. RNA binding RRF: ribosome release factor + 2 f. Met (f. Met-t. RNAf. Met) f. Met GTP 2 1 3 30 S Initiation Complex


30 S Initiation Complex 1 + f. Met GTP 2 1 3 f. Met 2 3 GTP 2 f. Met aa Psite Asite + GDP +Pi f. Met Elongation Phase of Translation 70 S Initiation Complex



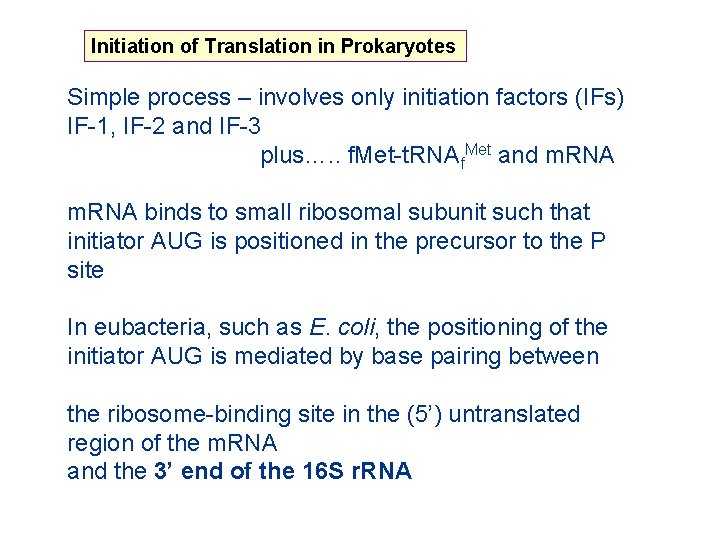
Initiation of Translation in Prokaryotes Simple process – involves only initiation factors (IFs) IF-1, IF-2 and IF-3 plus…. . f. Met-t. RNA f. Met and m. RNA binds to small ribosomal subunit such that initiator AUG is positioned in the precursor to the P site In eubacteria, such as E. coli, the positioning of the initiator AUG is mediated by base pairing between the ribosome-binding site in the (5’) untranslated region of the m. RNA and the 3’ end of the 16 S r. RNA

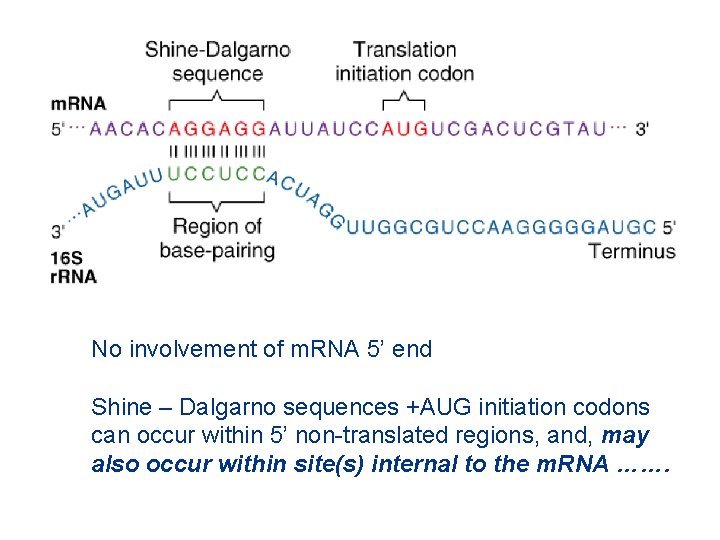
No involvement of m. RNA 5’ end Shine – Dalgarno sequences +AUG initiation codons can occur within 5’ non-translated regions, and, may also occur within site(s) internal to the m. RNA …….

Initiation of Translation in Prokaryotes

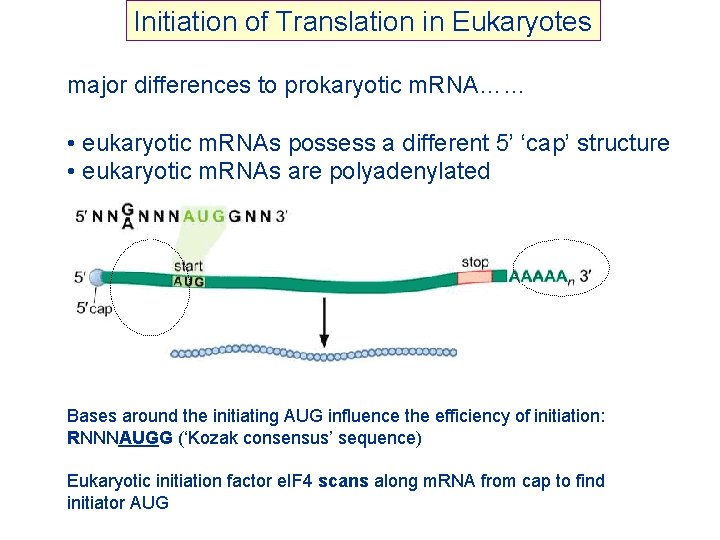
Initiation of Translation in Eukaryotes major differences to prokaryotic m. RNA…… • eukaryotic m. RNAs possess a different 5’ ‘cap’ structure • eukaryotic m. RNAs are polyadenylated Bases around the initiating AUG influence the efficiency of initiation: RNNNAUGG (‘Kozak consensus’ sequence) Eukaryotic initiation factor e. IF 4 scans along m. RNA from cap to find initiator AUG

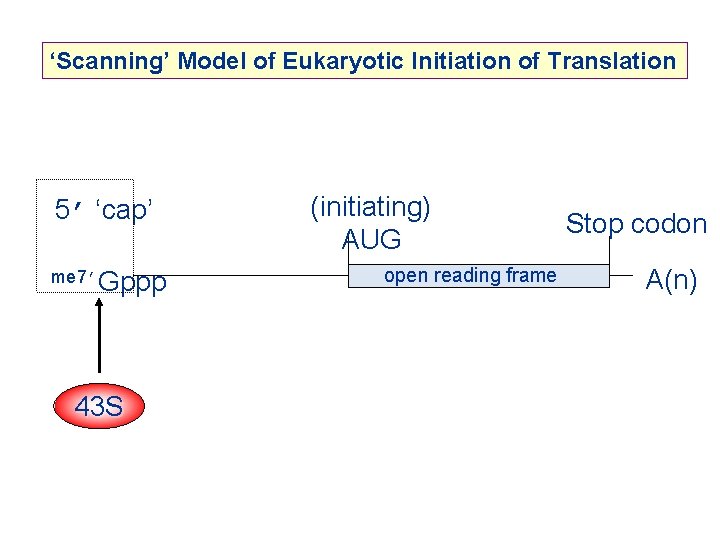
‘Scanning’ Model of Eukaryotic Initiation of Translation 5’ ‘cap’ me 7’Gppp 43 S (initiating) AUG open reading frame Stop codon A(n)

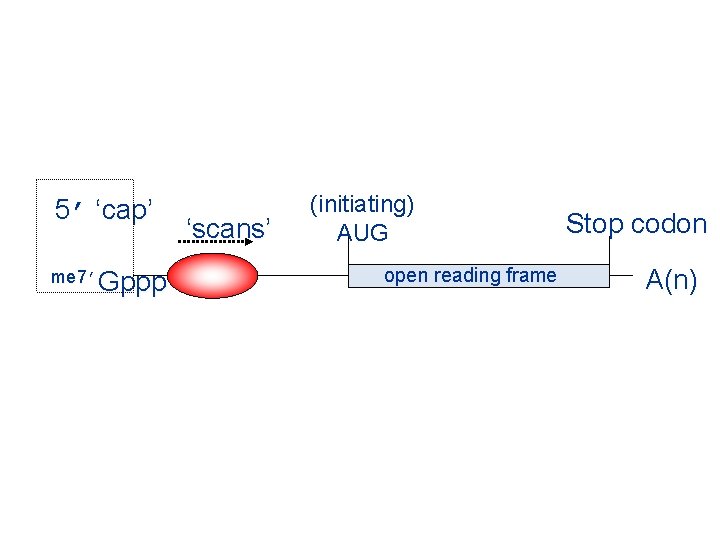
5’ ‘cap’ me 7’Gppp ‘scans’ (initiating) AUG open reading frame Stop codon A(n)

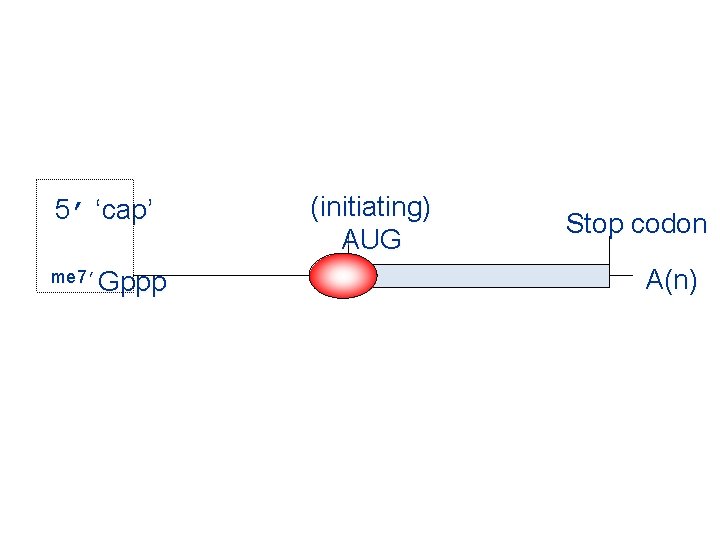
5’ ‘cap’ me 7’Gppp (initiating) AUG Stop codon A(n)

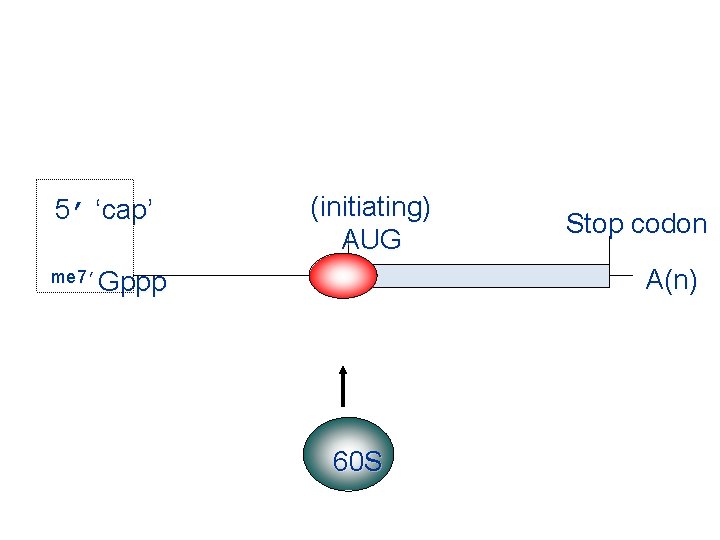
5’ ‘cap’ (initiating) AUG Stop codon A(n) me 7’Gppp 60 S

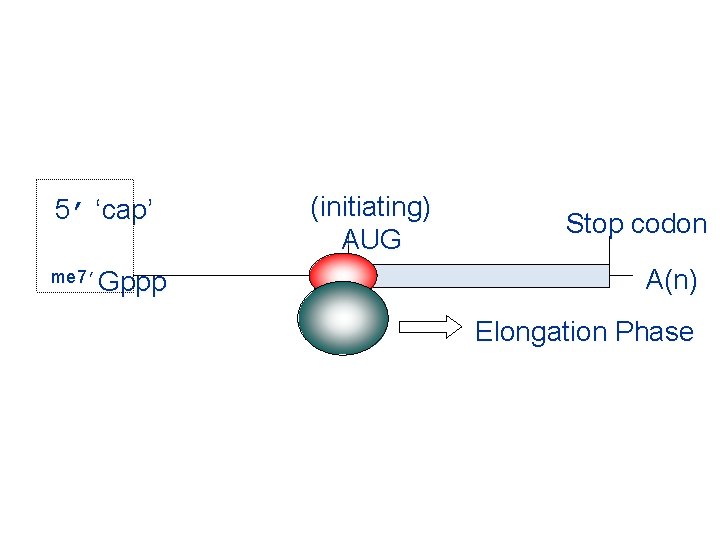
5’ ‘cap’ me 7’Gppp (initiating) AUG Stop codon A(n) Elongation Phase

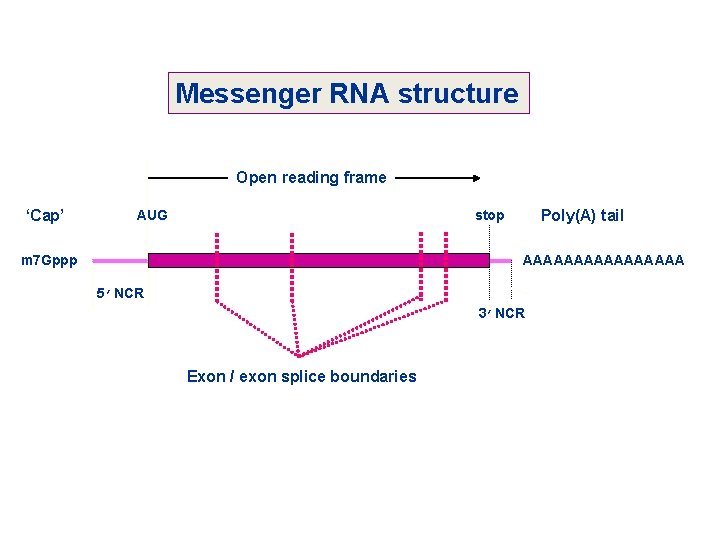
Messenger RNA structure Open reading frame ‘Cap’ AUG Poly(A) tail stop m 7 Gppp AAAAAAAA 5’NCR 3’NCR Exon / exon splice boundaries

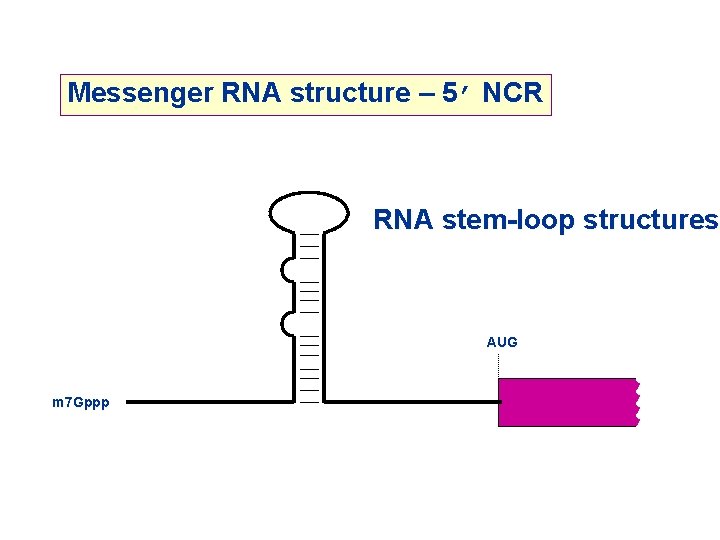
Messenger RNA structure – 5’ NCR RNA stem-loop structures AUG m 7 Gppp

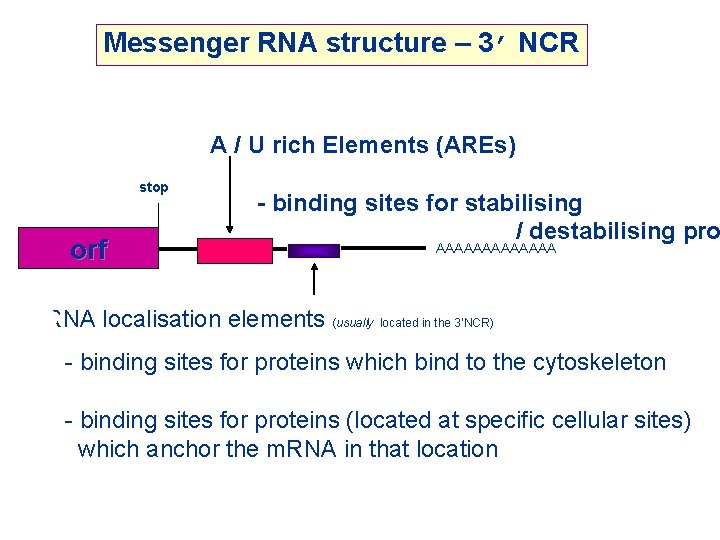
Messenger RNA structure – 3’ NCR A / U rich Elements (AREs) stop orf - binding sites for stabilising / destabilising pro AAAAAAA m. RNA localisation elements (usually located in the 3’NCR) - binding sites for proteins which bind to the cytoskeleton - binding sites for proteins (located at specific cellular sites) which anchor the m. RNA in that location






