Posttranscriptional Processes Processing of eukaryotic prem RNA capping




- Slides: 19

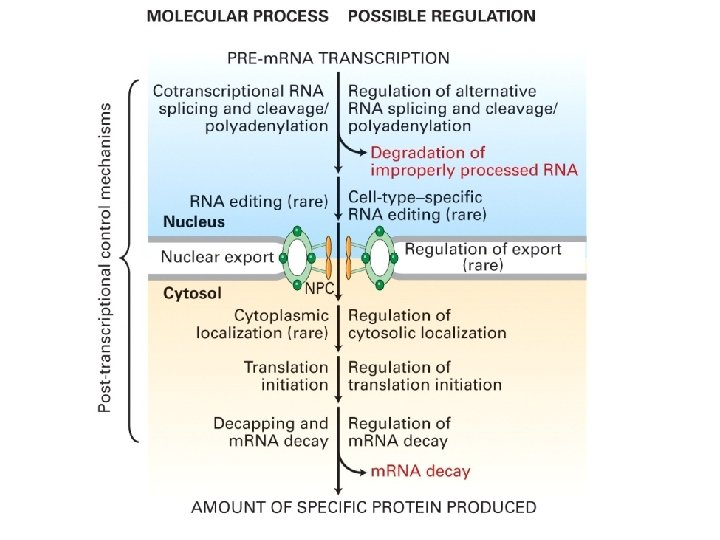
Post-transcriptional Processes

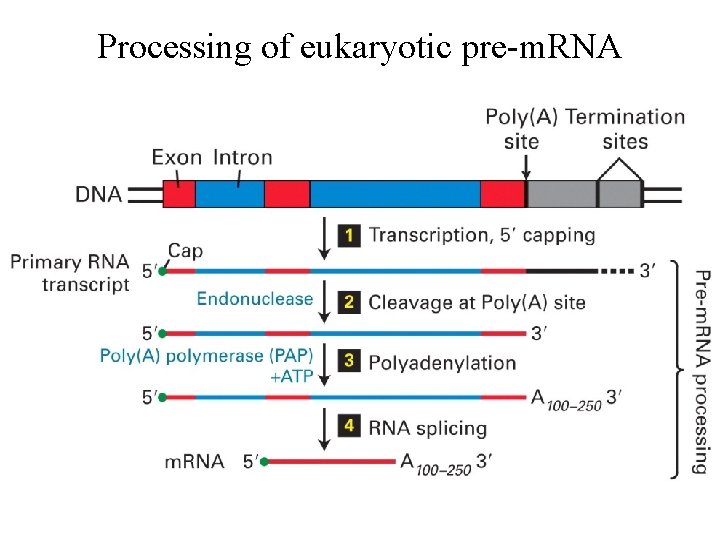
• Processing of eukaryotic pre-m. RNA -capping -polyadenylation -splicing -editing • Nuclear transport

RNA Processing • Very few RNA molecules are transcribed directly into the final mature RNA. • Most newly transcribed RNA molecules (primary transcripts) undergo various alterations to yield the mature product • RNA processing is the collective term used to describe the molecular events allowing the primary transcripts to become the mature RNA.


RNPs Ribonucleoproteins = RNA protein complexes Ø The RNA molecules in cells usually exist complexed with proteins Ø specific proteins attach to specific RNAs Ø Ribosomes are the largest and most complex RNPs

Ribozyme • Ribozymes are catalytic RNA molecules that can catalyze particular biochemical reactions. • RNase P RNA is a ribozyme. • RNase P RNA from bacteria is more catalytically active in vitro than those from eukaryotic and archaebacterial cells. All RNase P RNAs share common sequences and structures.

Self-splicing introns: the intervening RNA that catalyze the splicing of themselves from their precursor RNA, and the joining of the exon sequences 1. Group I introns, such as Tetrahymena intron 2. Group II introns. ¡

Processing of eukaryotic pre-m. RNA

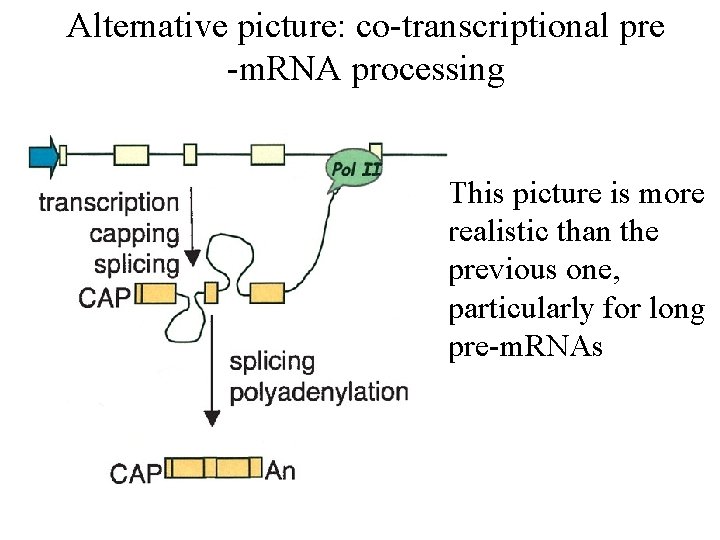
Alternative picture: co-transcriptional pre -m. RNA processing • This picture is more realistic than the previous one, particularly for long pre-m. RNAs

Heterogenous ribonucleoprotein patricles (hn. RNP) proteins • In nucleus nascent RNA transcripts are associated with abundant set of proteins • hn. RNPs prevent formation of secondary structures within pre-m. RNAs • hn. RNP proteins are multidomain with one or more RNA binding domains and at least one domain for interaction with other proteins • some hn. RNPs contribute to pre-m. RNA recognition by RNA processing enzymes

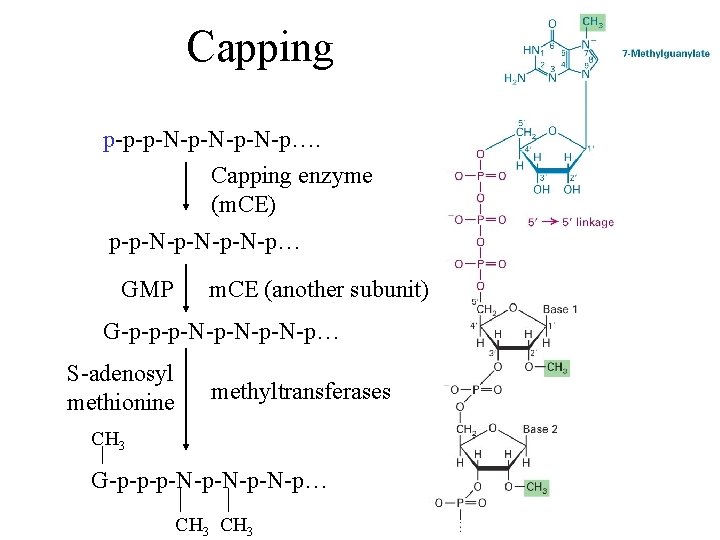
Capping p-p-p-N-p-N-p…. Capping enzyme (m. CE) p-p-N-p-N-p… GMP m. CE (another subunit) G-p-p-p-N-p-N-p… S-adenosyl methionine methyltransferases CH 3 G-p-p-p-N-p-N-p… CH 3

The capping enzyme • A bifunctional enzyme with both 5’-triphosphotase and guanyltransferase activities • In yeast the capping enzyme is a heterodimer • In metazoans the capping enzyme is monomeric with two catalytic domains • The capping enzyme specific only for RNAs, transcribed by RNA Pol II

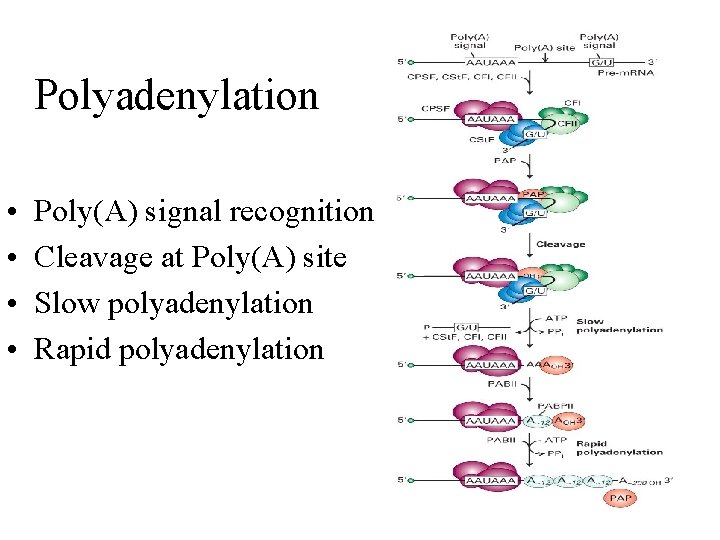
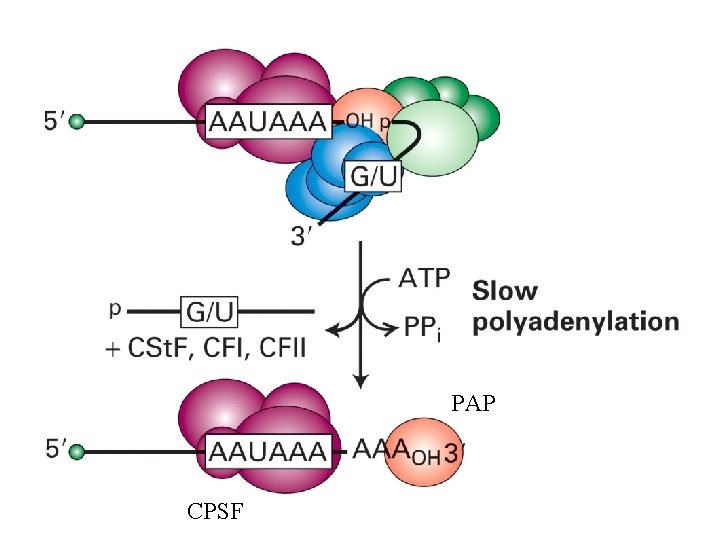
Polyadenylation • • Poly(A) signal recognition Cleavage at Poly(A) site Slow polyadenylation Rapid polyadenylation

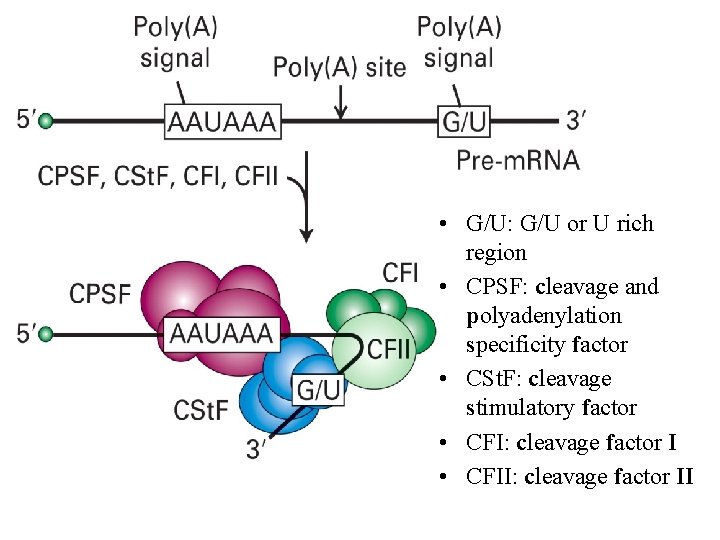
• G/U: G/U or U rich region • CPSF: cleavage and polyadenylation specificity factor • CSt. F: cleavage stimulatory factor • CFI: cleavage factor I • CFII: cleavage factor II

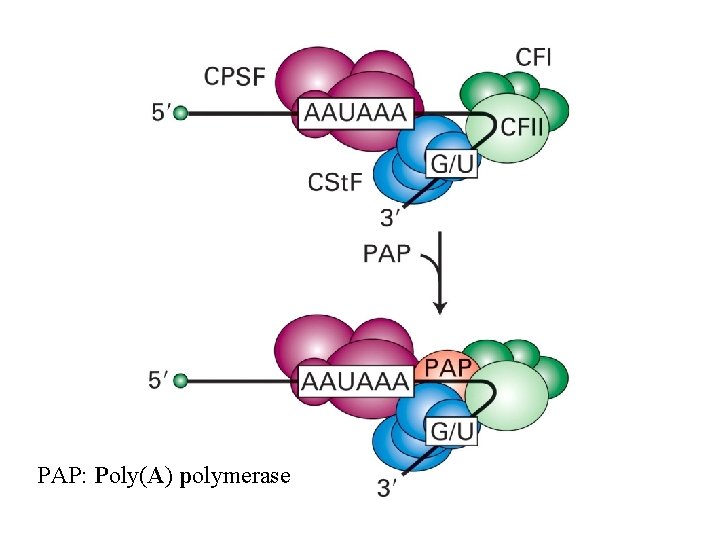
PAP: Poly(A) polymerase


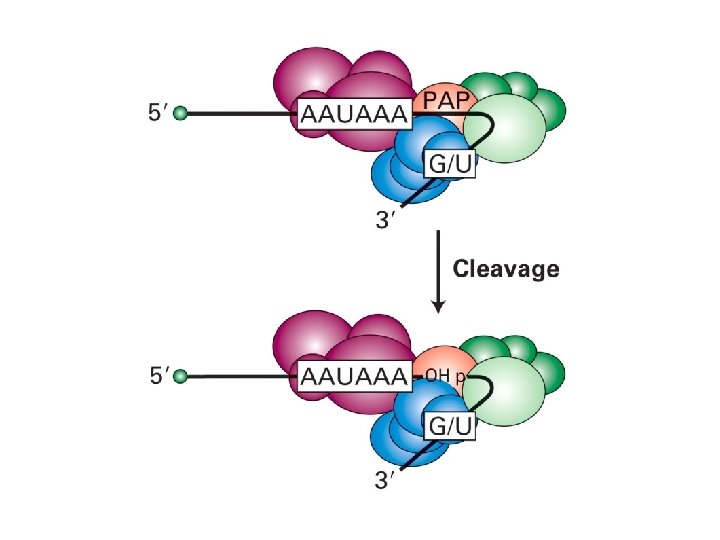
PAP CPSF

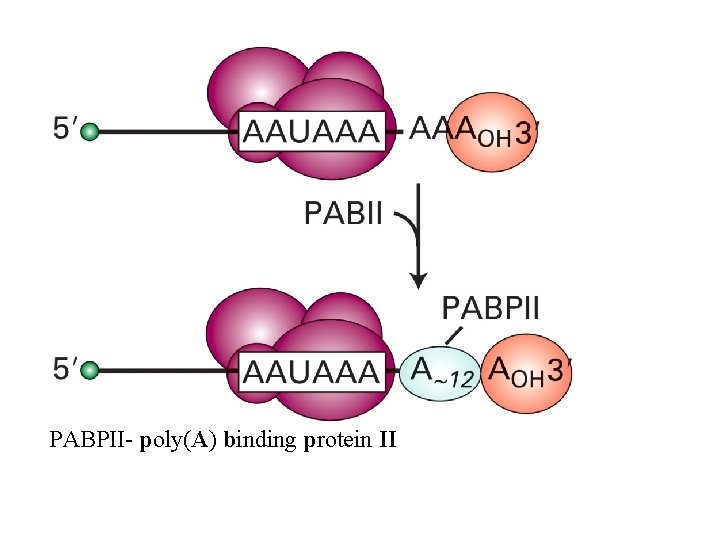
PABPII- poly(A) binding protein II

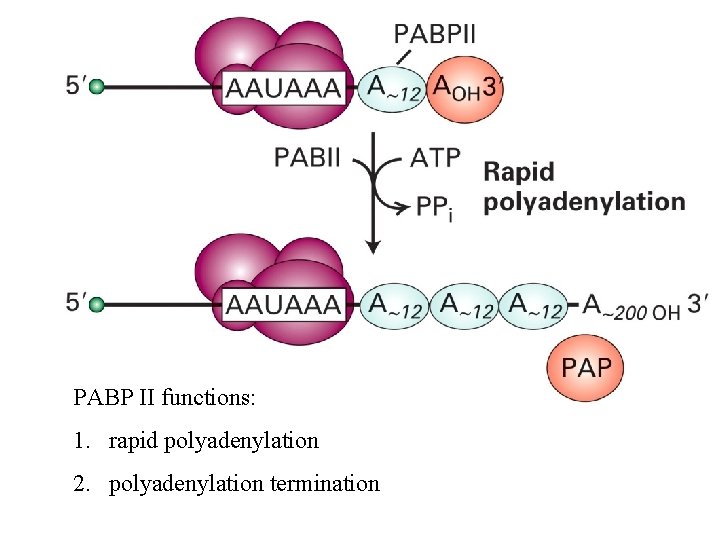
PABP II functions: 1. rapid polyadenylation 2. polyadenylation termination