RNA processing and Translation Eukaryotic cells modify RNA

- Slides: 19

RNA processing and Translation

Eukaryotic cells modify RNA after transcription (RNA processing) • During RNA processing, both ends of the primary transcript are usually altered • Also, usually some interior parts of the molecule are cut out, and the other parts spliced together Copyright © 2008 Pearson Education Inc. , publishing as Pearson Benjamin Cummings

Split Genes and RNA Splicing • Most eukaryotic genes and their RNA transcripts have long noncoding stretches of nucleotides that lie between coding regions • These noncoding regions are called intervening sequences, or introns • The other regions are called exons because they are eventually expressed, usually translated into amino acid sequences • RNA splicing removes introns and joins exons, creating an m. RNA molecule with a continuous coding sequence Copyright © 2008 Pearson Education Inc. , publishing as Pearson Benjamin Cummings

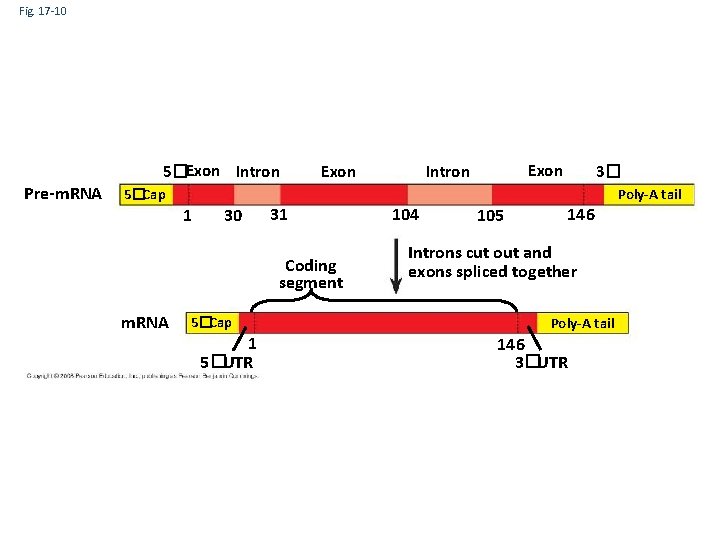
Fig. 17 -10 5�Exon Intron Pre-m. RNA Exon Intron Poly-A tail 5�Cap 1 30 31 Coding segment m. RNA 3� 5�Cap 1 5�UTR 104 105 146 Introns cut out and exons spliced together Poly-A tail 146 3�UTR

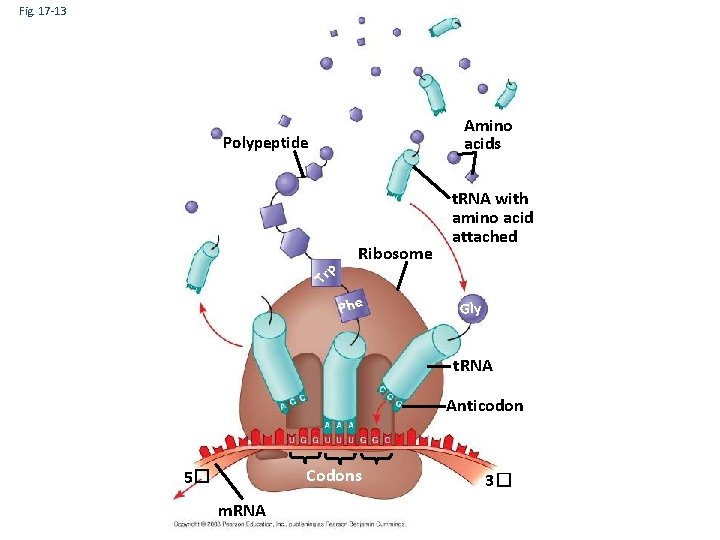
Molecular Components of Translation • A cell translates an m. RNA message into protein with the help of transfer RNA (t. RNA) • Molecules of t. RNA are not identical: – Each carries a specific amino acid on one end – Each has an anticodon on the other end; the anticodon base-pairs with a complementary codon on m. RNA Copyright © 2008 Pearson Education Inc. , publishing as Pearson Benjamin Cummings

Fig. 17 -13 Amino acids Polypeptide Tr p Ribosome t. RNA with amino acid attached Phe Gly t. RNA Anticodon Codons 5� m. RNA 3�

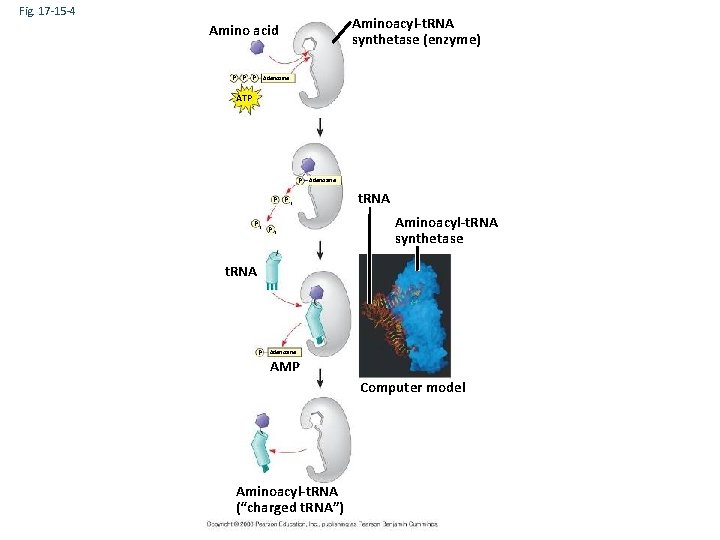
• Accurate translation requires two steps: – First: a correct match between a t. RNA and an amino acid, done by the enzyme aminoacyl-t. RNA synthetase – Second: a correct match between the t. RNA anticodon and an m. RNA codon Copyright © 2008 Pearson Education Inc. , publishing as Pearson Benjamin Cummings

Fig. 17 -15 -4 Aminoacyl-t. RNA synthetase (enzyme) Amino acid P P P Adenosine ATP P Adenosine P Pi Pi P i t. RNA Aminoacyl-t. RNA synthetase t. RNA P Adenosine AMP Computer model Aminoacyl-t. RNA (“charged t. RNA”)

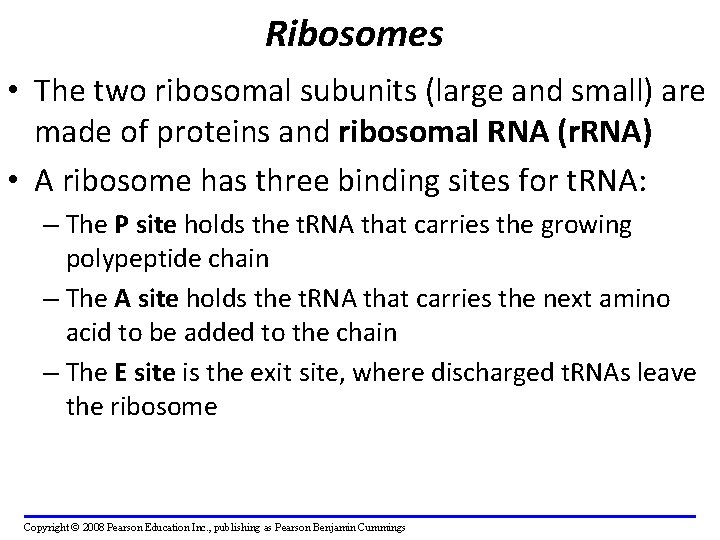
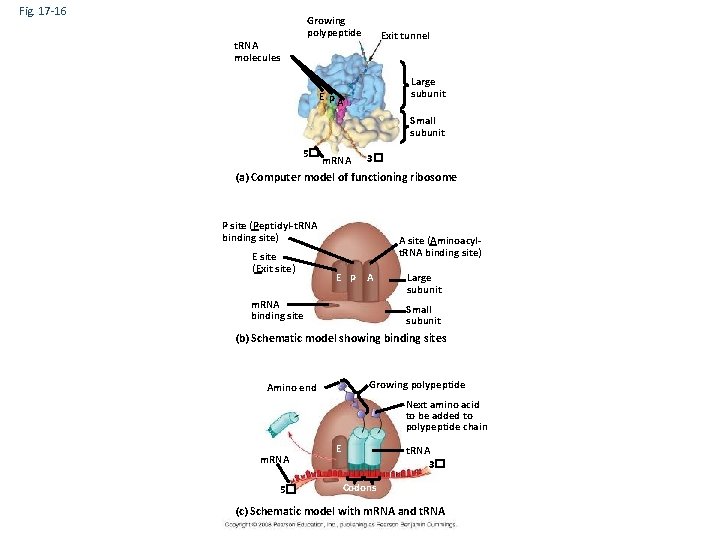
Ribosomes • The two ribosomal subunits (large and small) are made of proteins and ribosomal RNA (r. RNA) • A ribosome has three binding sites for t. RNA: – The P site holds the t. RNA that carries the growing polypeptide chain – The A site holds the t. RNA that carries the next amino acid to be added to the chain – The E site is the exit site, where discharged t. RNAs leave the ribosome Copyright © 2008 Pearson Education Inc. , publishing as Pearson Benjamin Cummings

Fig. 17 -16 Growing polypeptide t. RNA molecules Exit tunnel Large subunit EP A Small subunit 5� m. RNA 3� (a) Computer model of functioning ribosome P site (Peptidyl-t. RNA binding site) E site (Exit site) A site (Aminoacylt. RNA binding site) E P A m. RNA binding site Large subunit Small subunit (b) Schematic model showing binding sites Growing polypeptide Amino end Next amino acid to be added to polypeptide chain m. RNA 5� E t. RNA 3� Codons (c) Schematic model with m. RNA and t. RNA

Building a Polypeptide • The three stages of translation: – Initiation – Elongation – Termination Copyright © 2008 Pearson Education Inc. , publishing as Pearson Benjamin Cummings

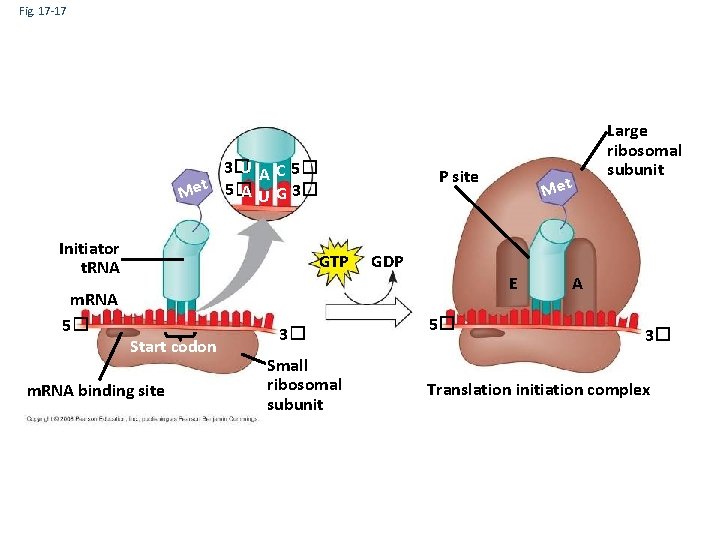
Ribosome Association and Initiation of Translation • The initiation stage of translation brings together m. RNA, a t. RNA with the first amino acid, and the two ribosomal subunits • First, a small ribosomal subunit binds with m. RNA and a special initiator t. RNA • Then the small subunit moves along the m. RNA until it reaches the start codon (AUG) • Proteins called initiation factors bring in the large subunit that completes the translation initiation complex Copyright © 2008 Pearson Education Inc. , publishing as Pearson Benjamin Cummings

Fig. 17 -17 3� U A C 5� A U G 3� Met 5� Initiator t. RNA m. RNA 5� P site GTP Start codon m. RNA binding site 3� Small ribosomal subunit Met Large ribosomal subunit GDP E 5� A 3� Translation initiation complex

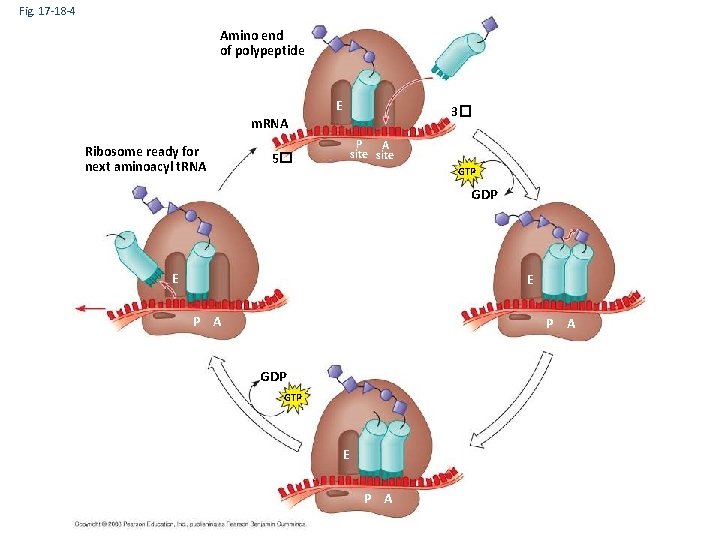
Elongation of the Polypeptide Chain • During the elongation stage, amino acids are added one by one to the preceding amino acid • Three steps: codon recognition, peptide bond formation, and translocation Copyright © 2008 Pearson Education Inc. , publishing as Pearson Benjamin Cummings

Fig. 17 -18 -4 Amino end of polypeptide E 3� m. RNA Ribosome ready for next aminoacyl t. RNA P A site 5� GTP GDP E E P A GDP GTP E P A

Termination of Translation • Termination occurs when a stop codon in the m. RNA reaches the A site of the ribosome • A water molecule is then added instead of an amino acid • This reaction releases the polypeptide, and the translation assembly then comes apart Translation Transcription/Translation Copyright © 2008 Pearson Education Inc. , publishing as Pearson Benjamin Cummings Transcription/Transation

Targeting Polypeptides to Specific Locations • Two populations of ribosomes are evident in cells: free ribsomes (in the cytosol) and bound ribosomes (attached to the ER) • Free ribosomes mostly synthesize proteins that function in the cytosol • Bound ribosomes make proteins of the endomembrane system and proteins that are secreted from the cell

Point mutations can affect protein structure and function • Mutations are changes in the genetic material of a cell or virus • Point mutations are chemical changes in just one base pair of a gene • Point mutations within a gene can be divided into two general categories – Base-pair substitutions – Base-pair insertions or deletions Copyright © 2008 Pearson Education Inc. , publishing as Pearson Benjamin Cummings

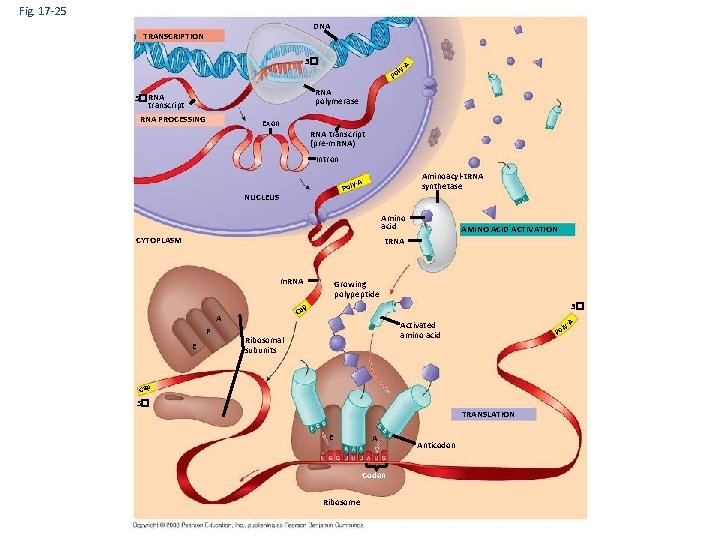
Fig. 17 -25 DNA TRANSCRIPTION 3� A ly- Po RNA polymerase 5�RNA transcript RNA PROCESSING Exon RNA transcript (pre-m. RNA) Intron Aminoacyl-t. RNA synthetase -A Poly NUCLEUS Amino acid CYTOPLASM AMINO ACID ACTIVATION t. RNA m. RNA E 3� p Ca A P Growing polypeptide Ribosomal subunits l Po Cap 5� TRANSLATION E A Codon Ribosome Anticodon A y- Activated amino acid