Eukaryotic m RNA Degradation Reading General m RNA





- Slides: 18

Eukaryotic m. RNA Degradation Reading: General m. RNA decay: Mitchell and Tollervey Current opinion in Genetics and Dev. 2000 10: 193 -198. NMD: Lykke-Andersen et al Science 2001 293: 1836 -1839. RNAi: Zamore Nature Structural Biology 2001 8 : 746 - 750.

outline Cis-acting m. RNA elements Pathways of decay Deadenylation Subsequent 5’-decay Subsequent 3’-decay Nonsense-mediated decay RNAi

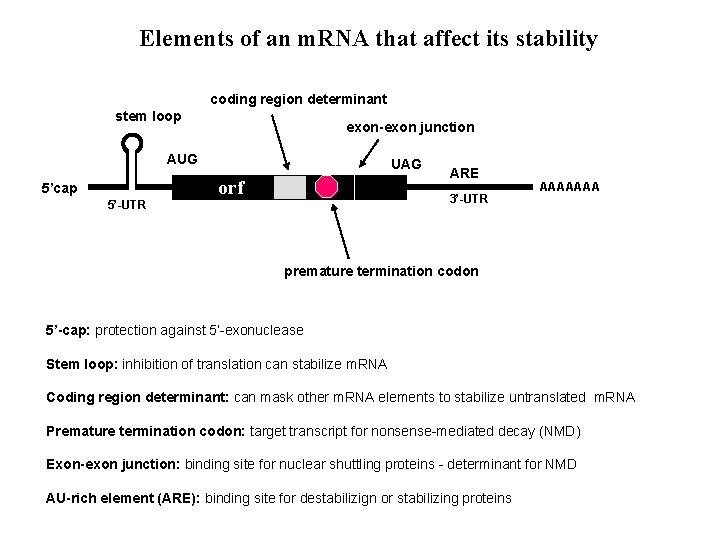
Elements of an m. RNA that affect its stability coding region determinant stem loop exon-exon junction AUG 5’cap 5’-UTR UAG orf ARE 3’-UTR AAAAAAA premature termination codon 5’-cap: protection against 5’-exonuclease Stem loop: inhibition of translation can stabilize m. RNA Coding region determinant: can mask other m. RNA elements to stabilize untranslated m. RNA Premature termination codon: target transcript for nonsense-mediated decay (NMD) Exon-exon junction: binding site for nuclear shuttling proteins - determinant for NMD AU-rich element (ARE): binding site for destabilizign or stabilizing proteins

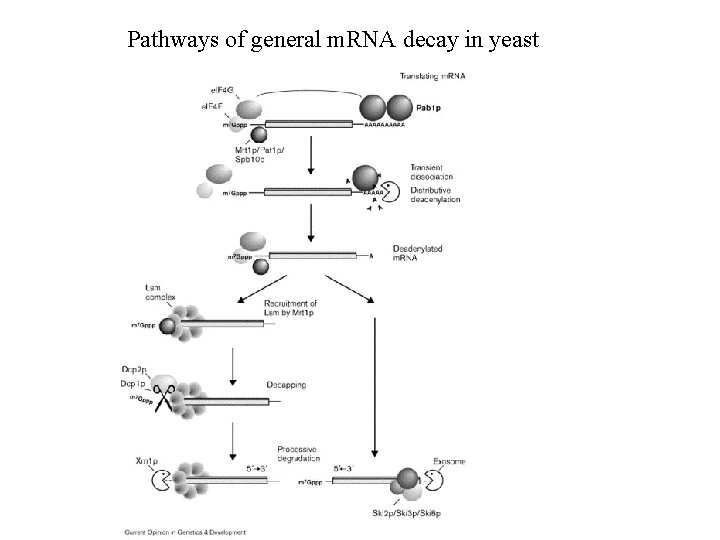
Pathways of general m. RNA decay in yeast

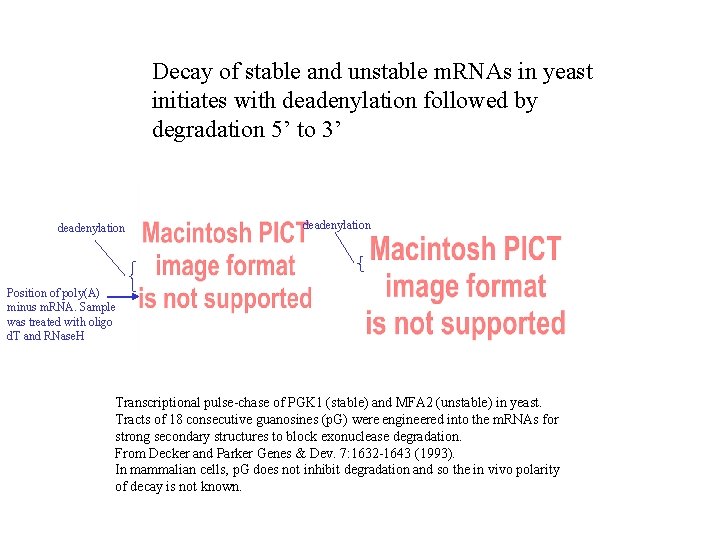
Decay of stable and unstable m. RNAs in yeast initiates with deadenylation followed by degradation 5’ to 3’ deadenylation Position of poly(A) minus m. RNA. Sample was treated with oligo d. T and RNase. H Transcriptional pulse-chase of PGK 1 (stable) and MFA 2 (unstable) in yeast. Tracts of 18 consecutive guanosines (p. G) were engineered into the m. RNAs for strong secondary structures to block exonuclease degradation. From Decker and Parker Genes & Dev. 7: 1632 -1643 (1993). In mammalian cells, p. G does not inhibit degradation and so the in vivo polarity of decay is not known.

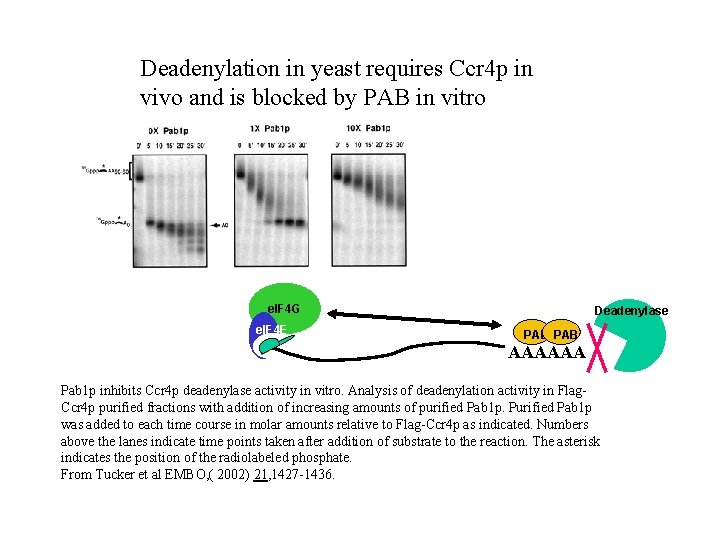
Deadenylation in yeast requires Ccr 4 p in vivo and is blocked by PAB in vitro e. IF 4 G e. IF 4 E Deadenylase PAB AAAAAA Pab 1 p inhibits Ccr 4 p deadenylase activity in vitro. Analysis of deadenylation activity in Flag. Ccr 4 p purified fractions with addition of increasing amounts of purified Pab 1 p. Purified Pab 1 p was added to each time course in molar amounts relative to Flag-Ccr 4 p as indicated. Numbers above the lanes indicate time points taken after addition of substrate to the reaction. The asterisk indicates the position of the radiolabeled phosphate. From Tucker et al EMBO, ( 2002) 21, 1427 -1436.

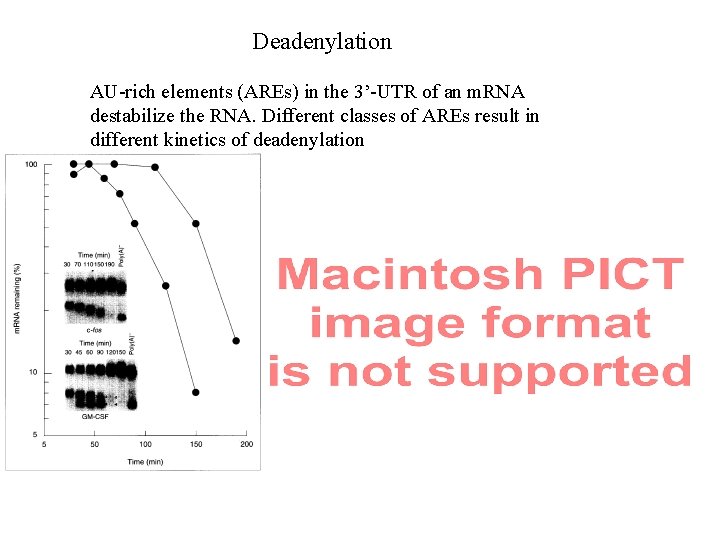
Deadenylation AU-rich elements (AREs) in the 3’-UTR of an m. RNA destabilize the RNA. Different classes of AREs result in different kinetics of deadenylation

Deadenylation in mammalian extracts requires DAN (or PARN) activity is stimulated by cap but access to the poly. A tail by DAN is blocked by the presence of translation factors as well as RNA binding proteins such as Hu. R. Other RNA binding proteins such as ARE-binding hn. RNP D destabilize m. RNAs possibly by disrupting PAB binding. Transient dissociation of PAB during translation or by the post termination ribosome may allow access for DAN. However, since deadenylation has not been recapitulated in extracts, the mechanism remains unclear. Also note that DAN is not related to yeast Ccr 4.

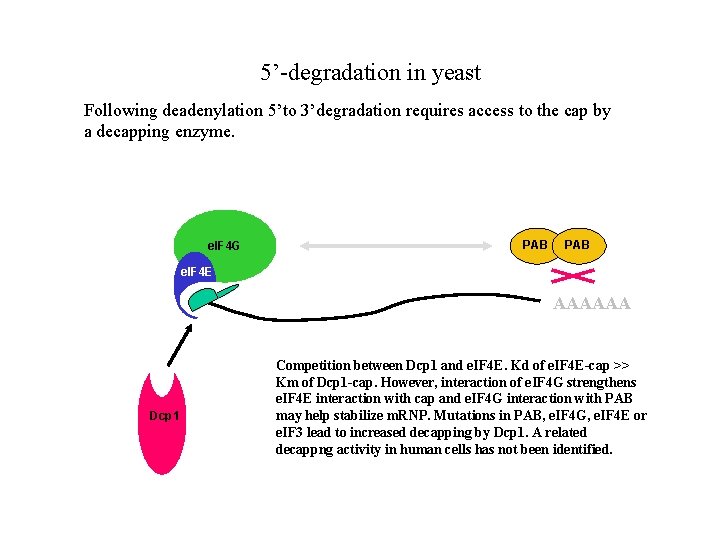
5’-degradation in yeast Following deadenylation 5’to 3’degradation requires access to the cap by a decapping enzyme. e. IF 4 G PAB e. IF 4 E AAAAAA Dcp 1 Competition between Dcp 1 and e. IF 4 E. Kd of e. IF 4 E-cap >> Km of Dcp 1 -cap. However, interaction of e. IF 4 G strengthens e. IF 4 E interaction with cap and e. IF 4 G interaction with PAB may help stabilize m. RNP. Mutations in PAB, e. IF 4 G, e. IF 4 E or e. IF 3 lead to increased decapping by Dcp 1. A related decappng activity in human cells has not been identified.

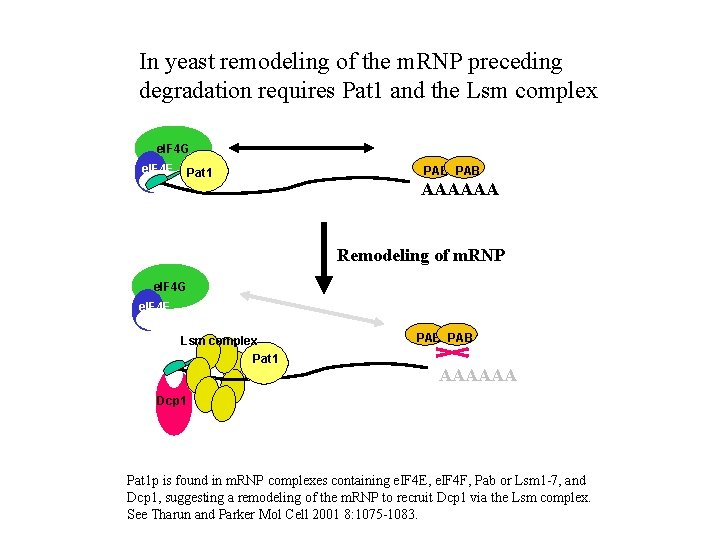
In yeast remodeling of the m. RNP preceding degradation requires Pat 1 and the Lsm complex e. IF 4 G e. IF 4 E Pat 1 PAB AAAAAA Remodeling of m. RNP e. IF 4 G e. IF 4 E Lsm complex Pat 1 PAB AAAAAA Dcp 1 Pat 1 p is found in m. RNP complexes containing e. IF 4 E, e. IF 4 F, Pab or Lsm 1 -7, and Dcp 1, suggesting a remodeling of the m. RNP to recruit Dcp 1 via the Lsm complex. See Tharun and Parker Mol Cell 2001 8: 1075 -1083.

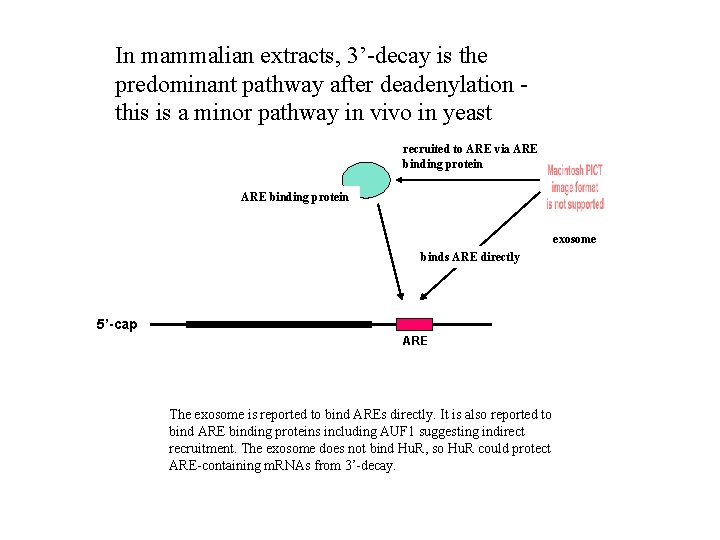
In mammalian extracts, 3’-decay is the predominant pathway after deadenylation this is a minor pathway in vivo in yeast recruited to ARE via ARE binding protein exosome binds ARE directly 5’-cap ARE The exosome is reported to bind AREs directly. It is also reported to bind ARE binding proteins including AUF 1 suggesting indirect recruitment. The exosome does not bind Hu. R, so Hu. R could protect ARE-containing m. RNAs from 3’-decay.

Purified exosome is inactive and apparently requires adapters (helicases) for substrate recognition Models for the activation of the exosome. a, Proteasome model. Access to the active sites of the exonucleases in the exosome is regulated by the RNA helicase Mtr 4 p or Ski 2 p (blue oval). Displacement or reorganization of the helicase occurs upon interaction with the RNP substrate. ATP hydrolysis by the activated RNA helicase unfolds the RNA structure and channels the free 3' end into the lumen of the exosome. b, Allosteric model. The RNA helicase interacts directly with the RNP substrate, recognizing specific marker proteins. The ATPase activity of the RNA helicase allows the exosome to be remodeled into an appropriately active form. The helicase may interact with the RNP substrate, either associated with the exosome or in its absence; for simplicity only the latter case is shown. From Mitchell Tollervey (2000) Nature Structural Biology 7, 843 – 846.

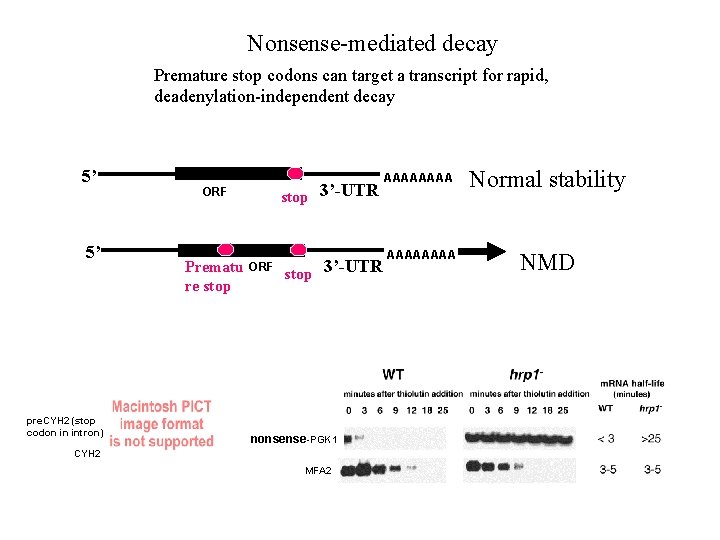
Nonsense-mediated decay Premature stop codons can target a transcript for rapid, deadenylation-independent decay 5’ 5’ pre. CYH 2 (stop codon in intron) stop 3’-UTR ORF Prematu re stop ORF stop 3’-UTR nonsense-PGK 1 CYH 2 MFA 2 AAAA Normal stability AAAA NMD

Model for NMD in mammalian cells From Lykke-Andersen Curr Biol. 2001 11(3): R 88 -91.

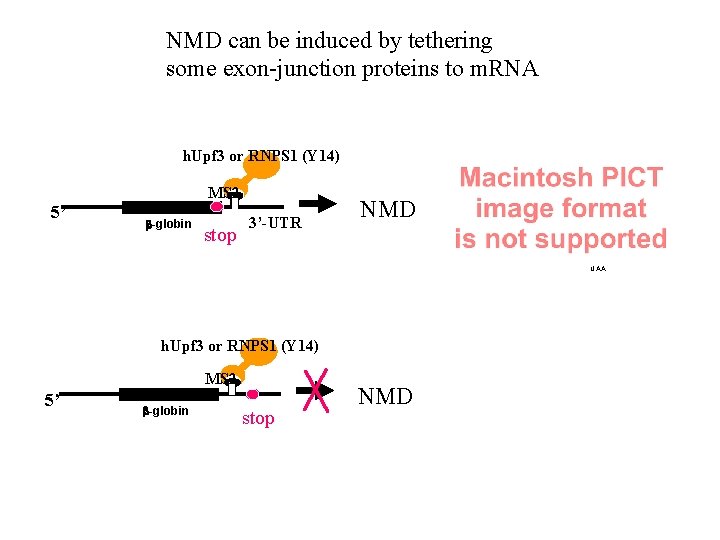
NMD can be induced by tethering some exon-junction proteins to m. RNA h. Upf 3 or RNPS 1 (Y 14) MS 2 5’ b-globin stop 3’-UTR NMD UAA h. Upf 3 or RNPS 1 (Y 14) MS 2 5’ b-globin stop NMD

In human cells cytoplasmic NMD may occur during first (pioneer) round of translation as a transcript is exported from the nucleus From Ishigaki et al Cell, 106: 607 -617 2001.

RNA interference (RNAi) From Zamore Nature Structural Biology 2001 8 : 746 - 750.

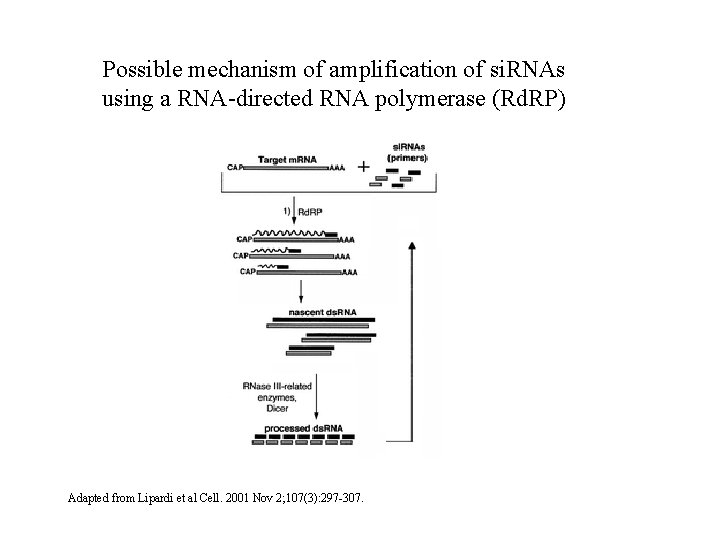
Possible mechanism of amplification of si. RNAs using a RNA-directed RNA polymerase (Rd. RP) Adapted from Lipardi et al Cell. 2001 Nov 2; 107(3): 297 -307.
 Pre reading while reading and post reading activities
Pre reading while reading and post reading activities Estimation of degradation function
Estimation of degradation function Purification table
Purification table Purines
Purines Mechanical degradation
Mechanical degradation Degradation of ketone bodies
Degradation of ketone bodies Importance of environmental degradation
Importance of environmental degradation Optimum notch filter in image processing
Optimum notch filter in image processing Light induced degradation
Light induced degradation Edman degradation
Edman degradation Salting out proteins
Salting out proteins Conclusion of environmental degradation
Conclusion of environmental degradation How environmental degradation occurs
How environmental degradation occurs Land degradation definition
Land degradation definition Sucrose synthesis
Sucrose synthesis Potential induced degradation
Potential induced degradation Abnormal degradation of disaccharides
Abnormal degradation of disaccharides Edman degradation
Edman degradation Tag degradation
Tag degradation