Pan India 1000 SARSCo V2 RNA Genome Sequencing


- Slides: 19

Pan India 1000 SARS-Co. V-2 RNA Genome Sequencing Consortium An Initiative of DBT-AIs Coordinated by National Institute of Biomedical Genomics (NIBMG), Kalyani Participating Institutes: Teams: NIBMG: Arindam Maitra, Nidhan Biswas, Saumitra Das ILS: Sunil Raghav, Arup Ghosh, Ajay Parida CDFD: Ashwin Dalal, Murali Bashyam, Debasis Mitra NCCS: Yogesh Souche, Manoj Kumar Bhat In. Stem-NCBS: Aswin Sai Narain, Dasaradhi Palakodepi, Satyajit Mayor, Apurva Sarin

The PAN–INDIA consortium launched the programme on April 27, 2020, to get the countrywide landscape of the variations in SARS-Co. V-2 RNA genome sequences. Coordinated by National Institute of Biomedical Genomics (NIBMG-Kalyani), West Bengal. Four other National clusters, ILS-Bhubaneswar, CDFD-Hyderabad, In. Stem-NCBS, Bangalore and NCCS-Pune have actively participated in sequencing and analysis. Other collaborating National Institutes and clinical organizations involved are ICMR-NICED, IPGMER-Kolkata, IISc. Bangalore, AIIMS-Uttarakhand, MAMC-Delhi, THSTI-Faridabad, GMC-Aurangabad, MGIMS-Wardha, RMRC -Bhubaneswar, AFMC and BJMC-Pune and other hospitals. Objectives: Short term: i) Sequencing of 1000 viral genomes from samples collected from different zones within India. ii) Molecular phylogeny of the virus, which will indicate the track of how the virus is evolving in India iii) Correlate sequence variation in viral RNA to disease severity and transmission efficiency, ultimately leading to identification of strains which are associated with enhanced pathogenicity. Long term: i) Identify host genetic polymorphisms that either confers susceptibility or protection from the viral infections. ii) Viral and host genetic determinants of disease severity

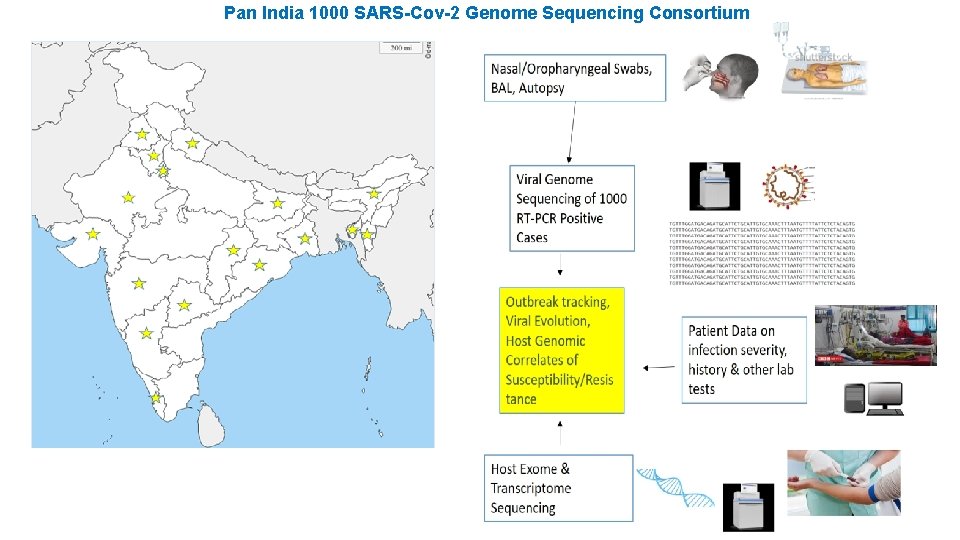
Pan India 1000 SARS-Cov-2 Genome Sequencing Consortium

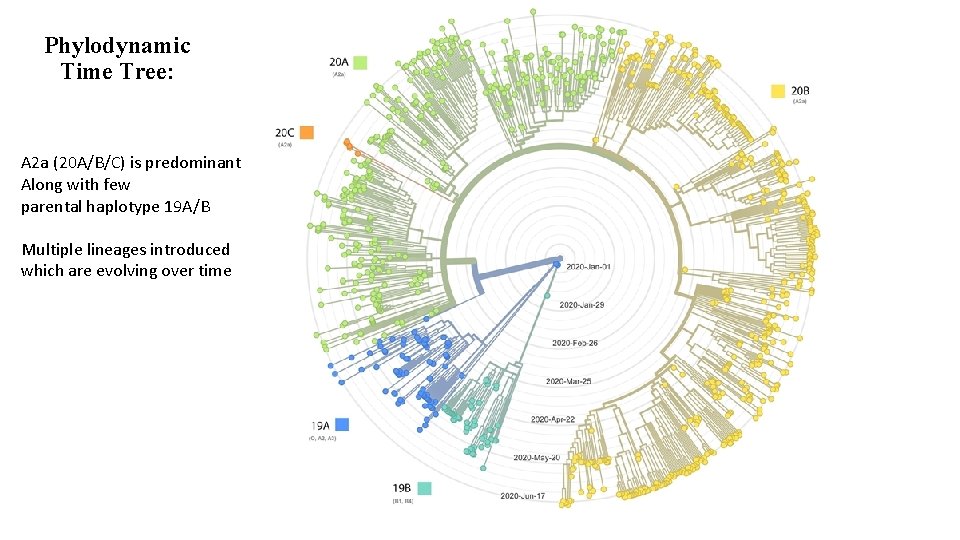
Phylodynamic Time Tree: A 2 a (20 A/B/C) is predominant Along with few parental haplotype 19 A/B Multiple lineages introduced which are evolving over time

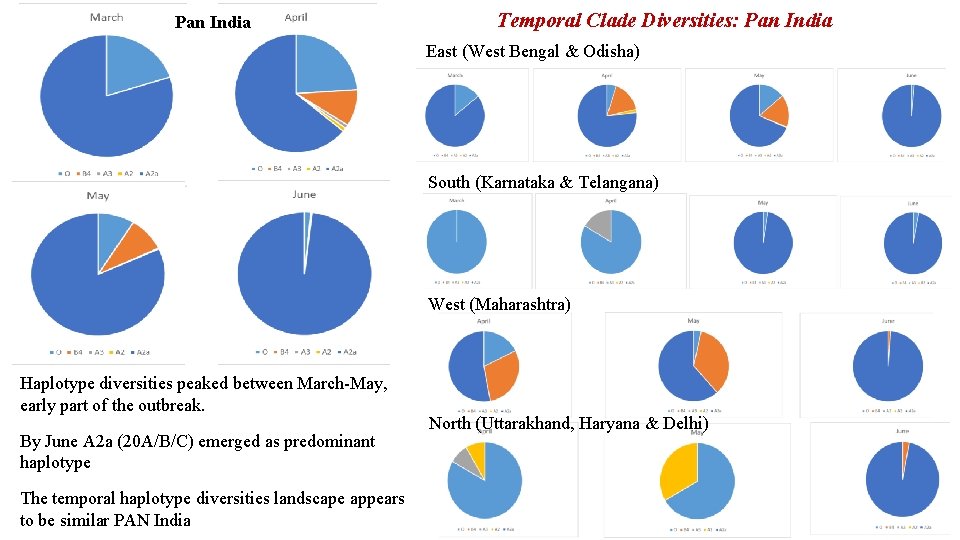
Pan India Temporal Clade Diversities: Pan India East (West Bengal & Odisha) South (Karnataka & Telangana) West (Maharashtra) Haplotype diversities peaked between March-May, early part of the outbreak. By June A 2 a (20 A/B/C) emerged as predominant haplotype The temporal haplotype diversities landscape appears to be similar PAN India North (Uttarakhand, Haryana & Delhi)

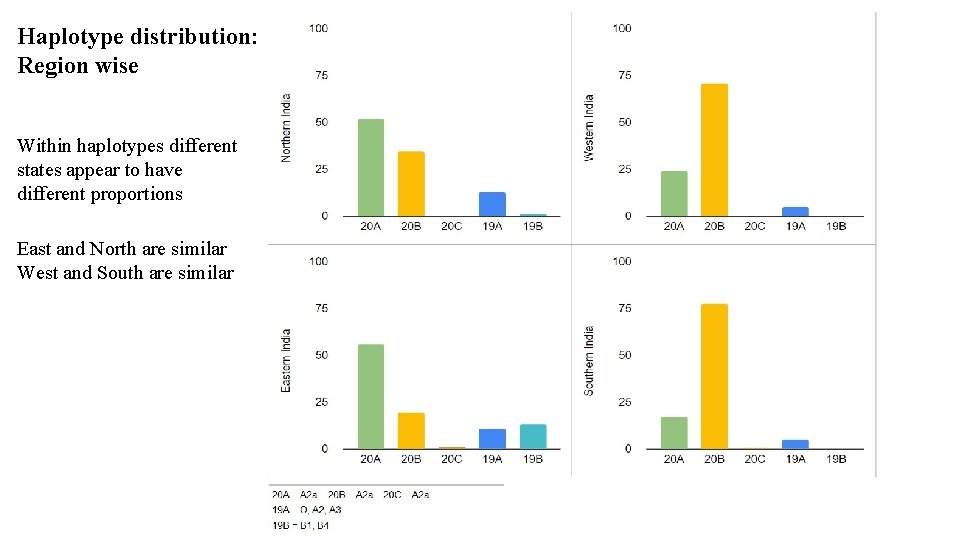
Haplotype distribution: Region wise Within haplotypes different states appear to have different proportions East and North are similar West and South are similar

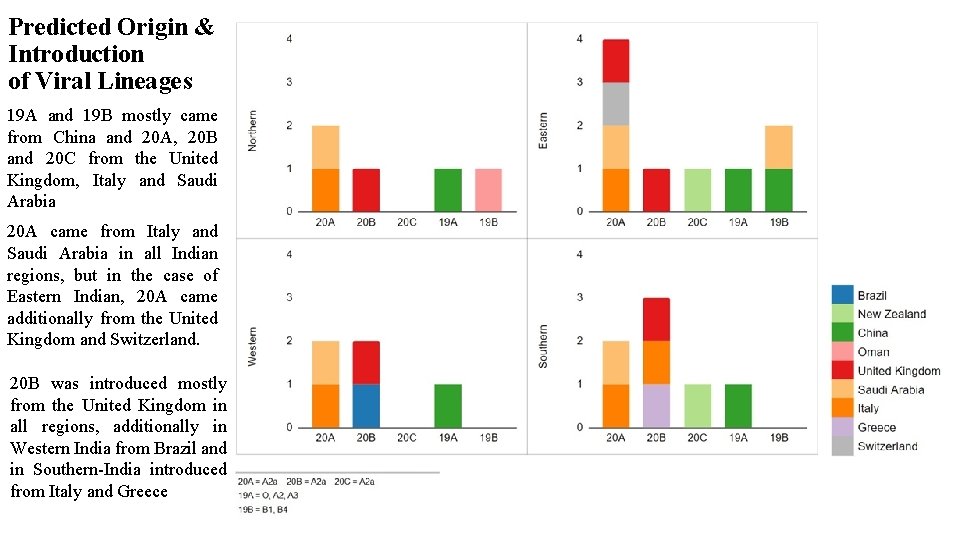
Predicted Origin & Introduction of Viral Lineages 19 A and 19 B mostly came from China and 20 A, 20 B and 20 C from the United Kingdom, Italy and Saudi Arabia 20 A came from Italy and Saudi Arabia in all Indian regions, but in the case of Eastern Indian, 20 A came additionally from the United Kingdom and Switzerland. 20 B was introduced mostly from the United Kingdom in all regions, additionally in Western India from Brazil and in Southern-India introduced from Italy and Greece

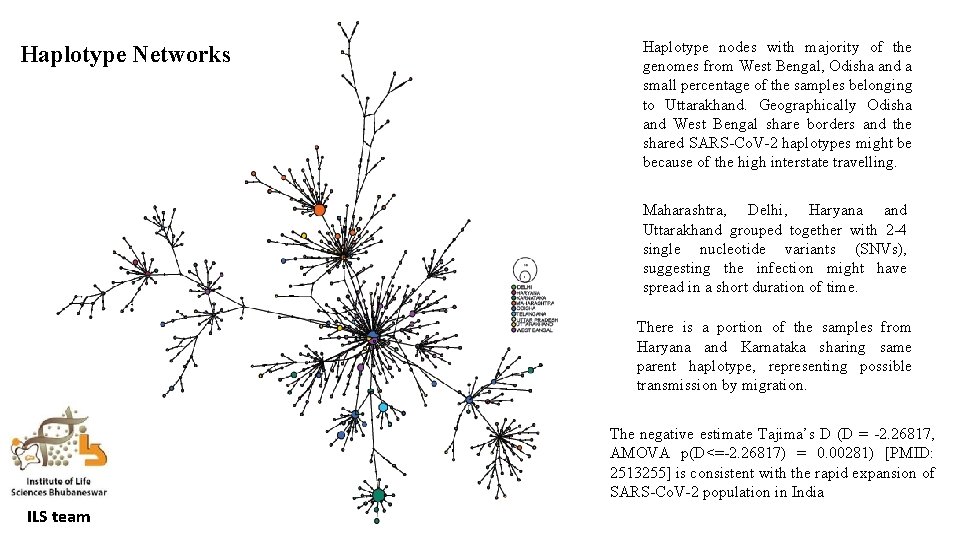
Haplotype Networks Haplotype nodes with majority of the genomes from West Bengal, Odisha and a small percentage of the samples belonging to Uttarakhand. Geographically Odisha and West Bengal share borders and the shared SARS-Co. V-2 haplotypes might be because of the high interstate travelling. Maharashtra, Delhi, Haryana and Uttarakhand grouped together with 2 -4 single nucleotide variants (SNVs), suggesting the infection might have spread in a short duration of time. There is a portion of the samples from Haryana and Karnataka sharing same parent haplotype, representing possible transmission by migration. The negative estimate Tajima’s D (D = -2. 26817, AMOVA p(D<=-2. 26817) = 0. 00281) [PMID: 2513255] is consistent with the rapid expansion of SARS-Co. V-2 population in India ILS team

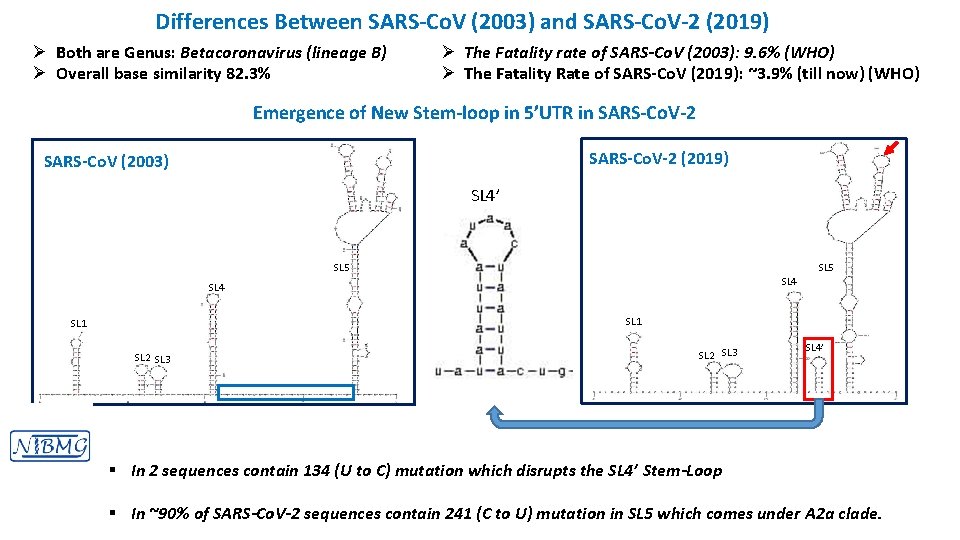
Differences Between SARS-Co. V (2003) and SARS-Co. V-2 (2019) Ø Both are Genus: Betacoronavirus (lineage B) Ø Overall base similarity 82. 3% Ø The Fatality rate of SARS-Co. V (2003): 9. 6% (WHO) Ø The Fatality Rate of SARS-Co. V (2019): ~3. 9% (till now) (WHO) Emergence of New Stem-loop in 5’UTR in SARS-Co. V-2 SARS-Co. V(2003) SARS-Co. V-2 (2019) SL 4’ SL 5 SL 4 SL 1 SL 2 SL 3 SL 4’ § In 2 sequences contain 134 (U to C) mutation which disrupts the SL 4’ Stem-Loop § In ~90% of SARS-Co. V-2 sequences contain 241 (C to U) mutation in SL 5 which comes under A 2 a clade.

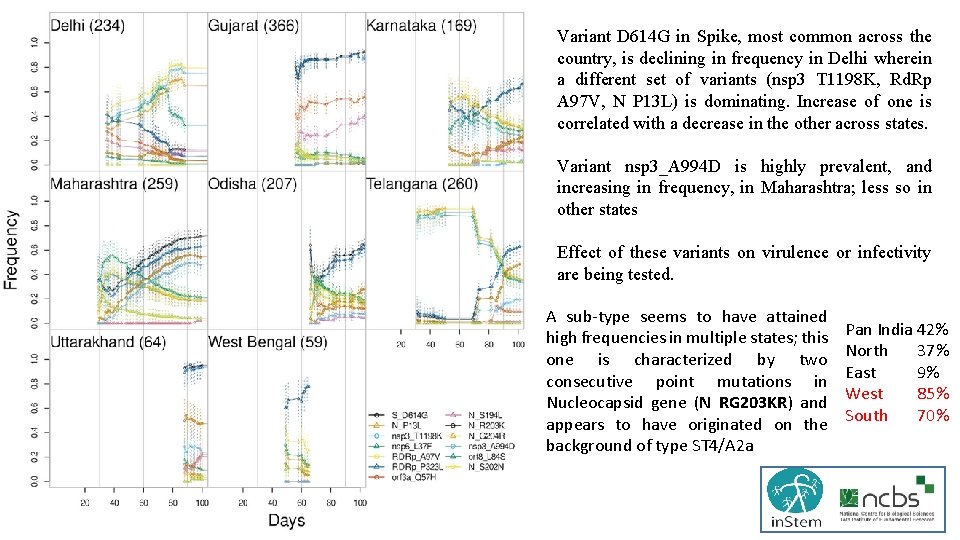
Variant D 614 G in Spike, most common across the country, is declining in frequency in Delhi wherein a different set of variants (nsp 3 T 1198 K, Rd. Rp A 97 V, N P 13 L) is dominating. Increase of one is correlated with a decrease in the other across states. Variant nsp 3_A 994 D is highly prevalent, and increasing in frequency, in Maharashtra; less so in other states Effect of these variants on virulence or infectivity are being tested. A sub-type seems to have attained high frequencies in multiple states; this one is characterized by two consecutive point mutations in Nucleocapsid gene (N RG 203 KR) and appears to have originated on the background of type ST 4/A 2 a Pan India 42% North 37% East 9% West 85% South 70%

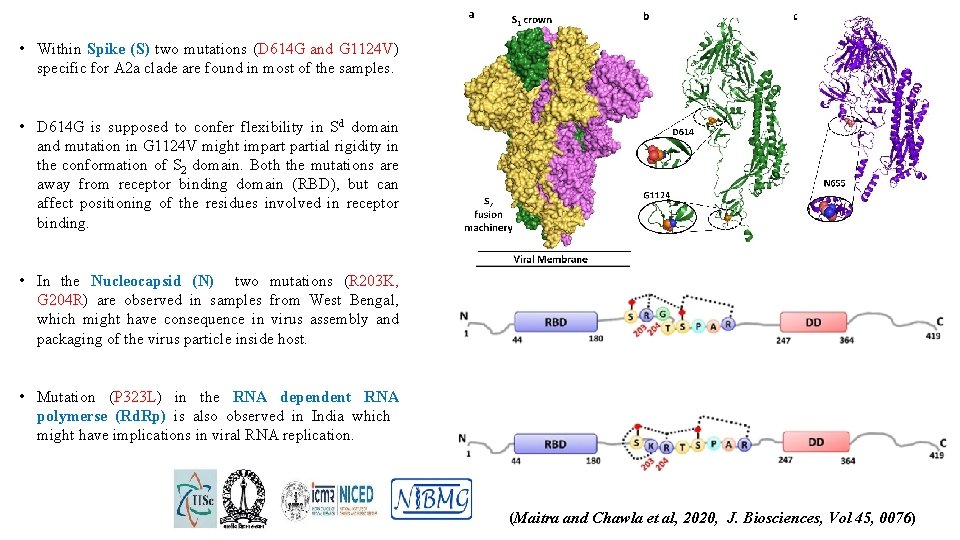
• Within Spike (S) two mutations (D 614 G and G 1124 V) specific for A 2 a clade are found in most of the samples. • D 614 G is supposed to confer flexibility in Sd domain and mutation in G 1124 V might impartial rigidity in the conformation of S 2 domain. Both the mutations are away from receptor binding domain (RBD), but can affect positioning of the residues involved in receptor binding. • In the Nucleocapsid (N) two mutations (R 203 K, G 204 R) are observed in samples from West Bengal, which might have consequence in virus assembly and packaging of the virus particle inside host. • Mutation (P 323 L) in the RNA dependent RNA polymerse (Rd. Rp) is also observed in India which might have implications in viral RNA replication. (Maitra and Chawla et al, 2020, J. Biosciences, Vol 45, 0076)

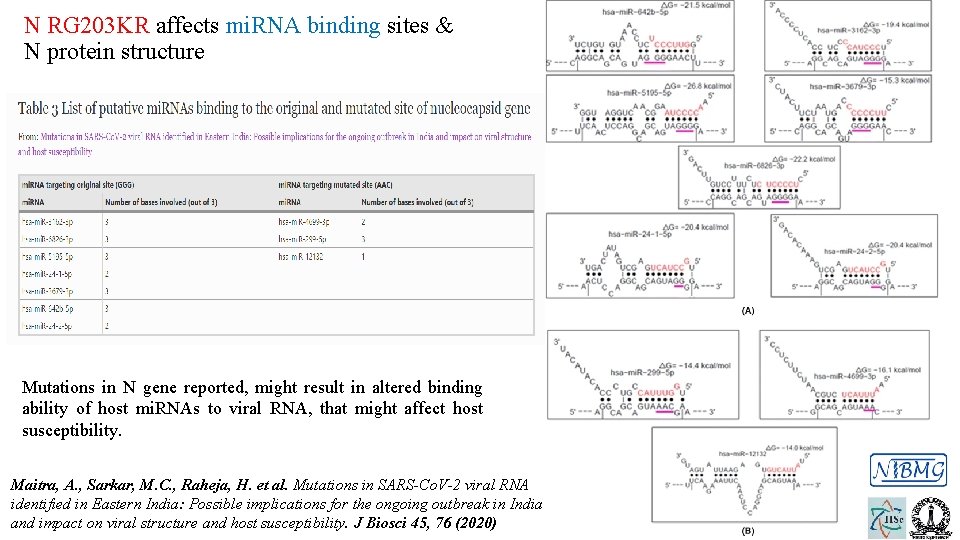
N RG 203 KR affects mi. RNA binding sites & N protein structure Mutations in N gene reported, might result in altered binding ability of host mi. RNAs to viral RNA, that might affect host susceptibility. Maitra, A. , Sarkar, M. C. , Raheja, H. et al. Mutations in SARS-Co. V-2 viral RNA identified in Eastern India: Possible implications for the ongoing outbreak in India and impact on viral structure and host susceptibility. J Biosci 45, 76 (2020)

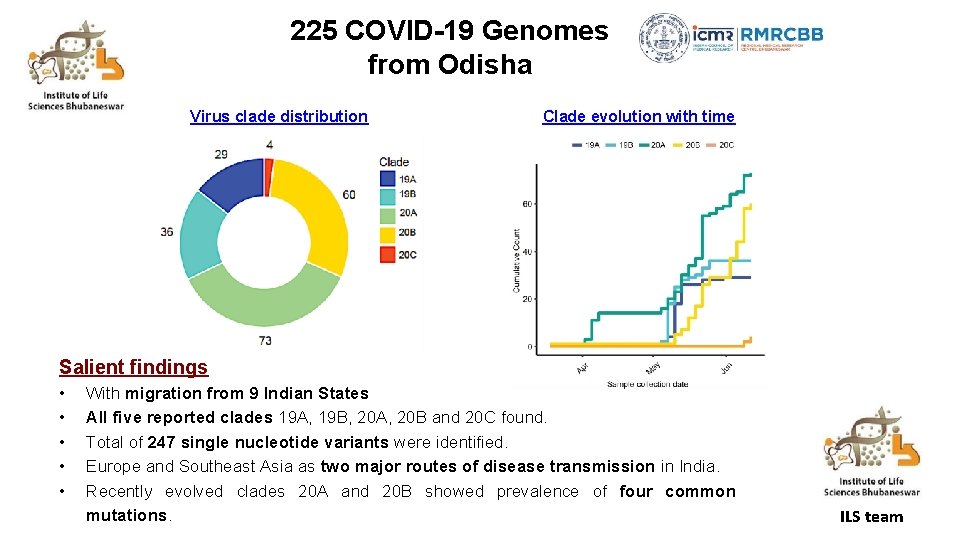
225 COVID-19 Genomes from Odisha Virus clade distribution Clade evolution with time Salient findings • • • With migration from 9 Indian States All five reported clades 19 A, 19 B, 20 A, 20 B and 20 C found. Total of 247 single nucleotide variants were identified. Europe and Southeast Asia as two major routes of disease transmission in India. Recently evolved clades 20 A and 20 B showed prevalence of four common mutations. ILS team

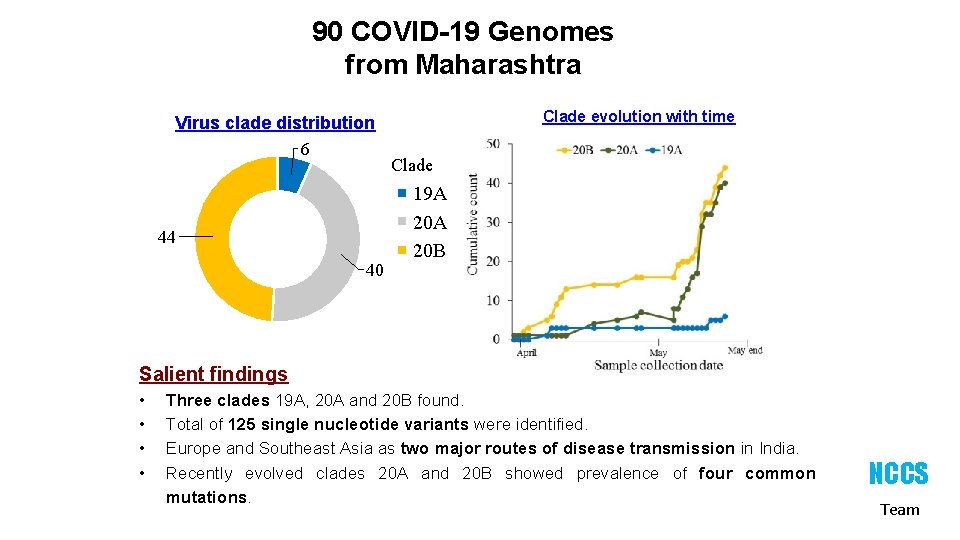
90 COVID-19 Genomes from Maharashtra Virus clade distribution 6 44 40 Clade evolution with time Clade 19 A 20 B Salient findings • • Three clades 19 A, 20 A and 20 B found. Total of 125 single nucleotide variants were identified. Europe and Southeast Asia as two major routes of disease transmission in India. Recently evolved clades 20 A and 20 B showed prevalence of four common mutations. NCCS Team

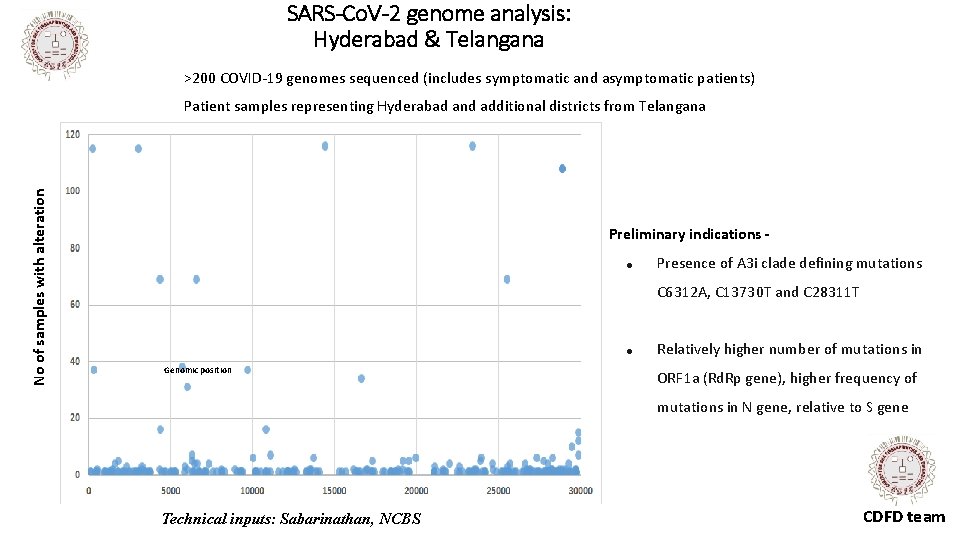
SARS-Co. V-2 genome analysis: Hyderabad & Telangana >200 COVID-19 genomes sequenced (includes symptomatic and asymptomatic patients) No of samples with alteration Patient samples representing Hyderabad and additional districts from Telangana Preliminary indications ● Presence of A 3 i clade defining mutations C 6312 A, C 13730 T and C 28311 T ● Genomic position Relatively higher number of mutations in ORF 1 a (Rd. Rp gene), higher frequency of mutations in N gene, relative to S gene Technical inputs: Sabarinathan, NCBS CDFD team

Highlights: As on date, the Consortium has achieved its initial goal of completing the sequencing of 1000 SARS-Co. V-2 genomes from nasopharyngeal and oropharyngeal swabs collected from individuals testing positive for COVID 19 by Real Time PCR. The samples were collected across 10 states covering different zones within India. Given the importance of this information for public health response initiatives investigating transmission of COVID-19, the sequence data will soon be released in public domain (GISAID database). Information will improve our understanding on how the virus is spreading, ultimately helping to interrupt the transmission chains, prevent new cases of infection, and provide impetus to research on intervention measures. Initial results indicate that multiple lineages of SARS-Co. V-2 are circulating in India, probably introduced by travel from Europe, USA and East Asia. In particular, there is a predominance of the A 2 a haplotype (20 A/B/C) with D 614 G mutation, which is found to be emerging in almost all regions of the country. This particular haplotype is globally reported to be associated with enhanced transmission efficiency. Additionally, mutations in important regions of the viral genome with significant geographical clustering have also been observed. Detailed mutational analysis to understand the gradual emergence of mutants at different regions of the country and its possible impact on the disease management is in progress.

Experimental & Computational Team NIBMG, Kalyani Dr. Arindam Maitra, Dr. Nidhan K. Biswas, Dr. Sreedhar Chinnaswamy, Mr. Shekhar Ghosh, Mr. Sumanta Sarkar, Dr. Subrata Patra, Dr. Rajib Mondal, Dr. Trinath Ghosh, Mr. Arnab Ghosh, Mr. Shouvik Chakraborty, Dr. Saumitra Das, ILS, Bhuabneswar Dr. Sunil Raghav. Mr. Arup Ghosh and Dr. Ajay Parida In. Stem-NCBS, Bengaluru Dr. Apurva Sarin, Dasaradhi Palakodeti, Aswin Sai Narain Seshasayee, Uma Ramakrishnan, Shah-e-Jahan Gulzar, Varadharajan Sundaramurthy, Srikar Krishna, Vanessa Molin Paynter, Awadhesh Pandit, Farhan Ali, Mohak Sharda, Dr. Satyajit Mayor, Dr. Apurva Sarin CDFD, Hyderabad Dr Ashwin Dalal, Dr Murali Bashyam, Dr Pratyusha Bala, Mr Vinay Donipadi, Dr Divya Vashisht, Dr Debashis Mitra, NCCS, Pune Dhiraj Paul, Kunal Jani, Janesh Kumar, Radha Chauhan, Vasudevan Seshadri, Girdhari Lal, Dr. Arvind Sahu, Dr. Yogesh S Shouche, Dr. Manoj Kumar Bhat, Coordinated by NIBMG, Kalyani

Clinical Collaborators ICMR-NICED Dr. Shanta Dutta, Dr. Mamta Chawla Sarkar, Ms. Ananya Chatterjee, Ms. Hasina Banu, Mr. Agniva Majumdar IPGMER, Kolkata Dr. Monimoy Banerjee, Dr. Raja Ray, Dr. Jayeeta Halder, Dr. Aritra Biswas GMC, Aurangabad Dr. Jyoti Iravan, Mr Dhaval Khatri, Mr Maitrik Dave IISc, Bengaluru Dr. Bharath K Sundararaj, Harsha Raheja, Prof. N. Srinivasan, Prof. Deepak K Saini, Prof. Amit Singh, Prof. K N Balaji, Prof. Umesh Varshney AIIMS, Rishikesh Prof Ravi Kant, Dr Deepjyoti Kalita, Dr Amit Mangla NIMS, Hyderabad Dr Madhumohan Rao, Dr Vijay Dharma Teja THSTI, Faridabad Dr. Guruprasad Medigeshi, Dr. Gagandeep Kang, Sharanabasava Patil, Anbalagan Ananthraj, Madhu Pareek, Imran Khan, ESIC Hospital and Medical College, Faridabad, Gurugram Civil Hospital, Gurugram and Palwal Civil Hospital, Palwal RMRC, Bhubaneswar Dr. Sanghamitra Pari, Dr. Jyotirmayee Turuk MAMC. Delhi Dr Sonal Saxena, Dr Vikas Manchanda, Dr Oves Siddiqui AFMC, Pune Sourav Sen, Santosh Karade, Kavita. Bala Anand, Shelinder Pal Singh Shergill, Rajiv Mohan Gupta BJMC, Pune Rajesh Karyakarte, Suvarna Joshi, Murlidhar Tambe MGIMS, Wardha Dr Vijayshri Deotale, Dr Rahul Narang, Dr Deepashri Maraskolhe

 Semi-global alignment
Semi-global alignment Perpartes
Perpartes Genome sequencing
Genome sequencing Hierarchical shotgun sequencing vs whole genome
Hierarchical shotgun sequencing vs whole genome Human genome project
Human genome project Shotgun sequencing
Shotgun sequencing Dna
Dna Sam file
Sam file 1000 genome project
1000 genome project Alternate splicing
Alternate splicing Vhf kurs gratis
Vhf kurs gratis Genome mapping
Genome mapping Act artemis
Act artemis Genome identification
Genome identification Chapter 14 the human genome
Chapter 14 the human genome Genome klick
Genome klick Repeated sequences
Repeated sequences Genome definition
Genome definition Genome.gov
Genome.gov Plant genome research program
Plant genome research program