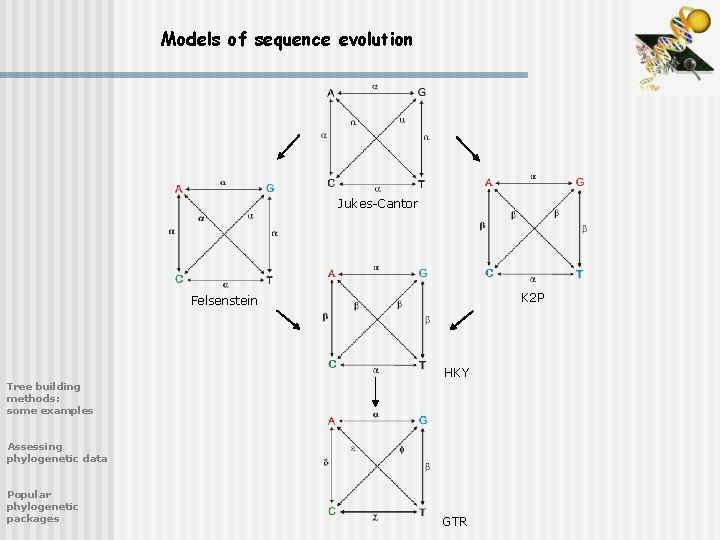
Models of sequence evolution JukesCantor K 2 P

- Slides: 24

Models of sequence evolution Jukes-Cantor K 2 P Felsenstein HKY Tree building methods: some examples Assessing phylogenetic data Popular phylogenetic packages GTR

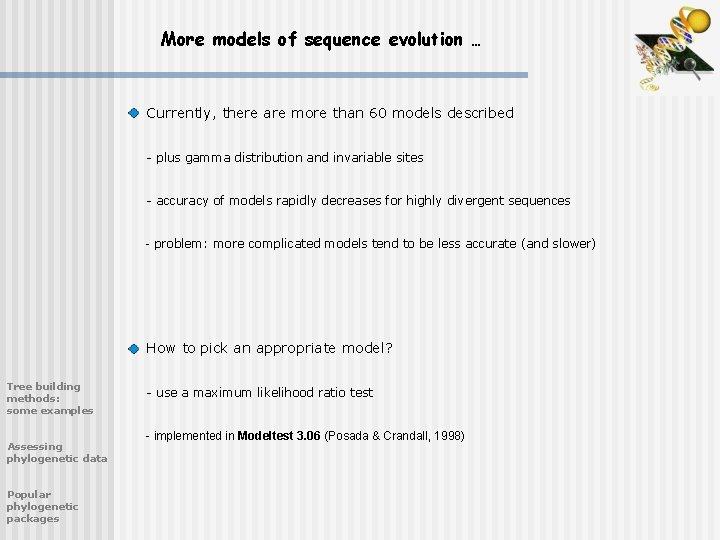
More models of sequence evolution … Currently, there are more than 60 models described - plus gamma distribution and invariable sites - accuracy of models rapidly decreases for highly divergent sequences - problem: more complicated models tend to be less accurate (and slower) How to pick an appropriate model? Tree building methods: some examples Assessing phylogenetic data Popular phylogenetic packages - use a maximum likelihood ratio test - implemented in Modeltest 3. 06 (Posada & Crandall, 1998)

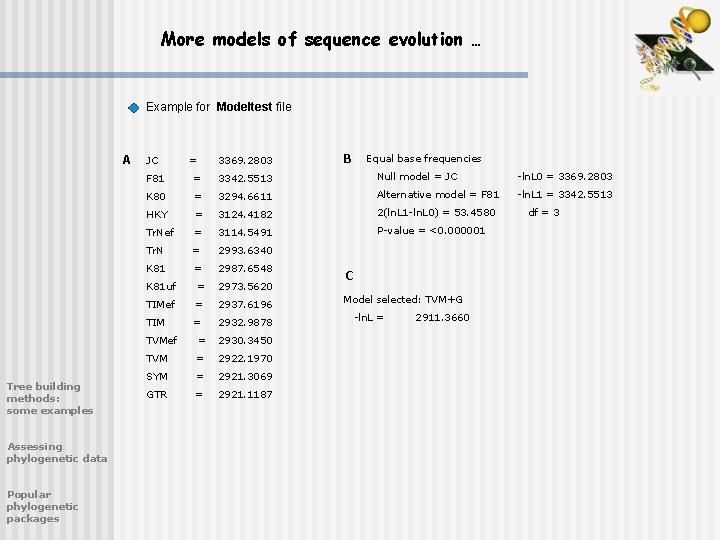
More models of sequence evolution … Example for Modeltest file A Tree building methods: some examples Assessing phylogenetic data Popular phylogenetic packages JC = 3369. 2803 B Equal base frequencies F 81 = 3342. 5513 Null model = JC -ln. L 0 = 3369. 2803 K 80 = 3294. 6611 Alternative model = F 81 -ln. L 1 = 3342. 5513 HKY = 3124. 4182 2(ln. L 1 -ln. L 0) = 53. 4580 Tr. Nef = 3114. 5491 P-value = <0. 000001 Tr. N = 2993. 6340 K 81 = 2987. 6548 K 81 uf = 2973. 5620 TIMef = 2937. 6196 TIM = 2932. 9878 TVMef = 2930. 3450 TVM = 2922. 1970 SYM = 2921. 3069 GTR = 2921. 1187 C Model selected: TVM+G -ln. L = 2911. 3660 df = 3

More models of sequence evolution … Amino acid sequences - infinitely more complicated than nucleotide sequences - some amino acids can replace one another with relatively little effect on the structure and function of the final protein while other replacements can be functionally devastating - from the standpoint of the genetic code, some amino acid changes can be made by a single DNA mutation while others require two or even three changes in the DNA sequence - in practice, what has been done is to calculate tables of frequencies of all amino acid replacements within families of related protein sequences in the databanks: i. e. PAM and BLOSSUM Tree building methods: some examples Assessing phylogenetic data Popular phylogenetic packages

Phylogenetic Inference II Disclaimers Before describing any theoretical or practical aspects of phylogenetics, it is necessary to give some disclaimers. This area of computational biology is an intellectual minefield! Neither theory nor the practical applications of any algorithms are universally accepted throughout the scientific community. The application of different software packages to a data set is very likely to give different answers; minor changes to a data set are also likely to profoundly change the result. Tree building methods: some examples Assessing phylogenetic data Popular phylogenetic packages

CS 177 Phylogenetics II Tree building methods: some examples Assessing phylogenetic data Popular phylogenetic software packages Tree building methods: some examples Assessing phylogenetic data Popular phylogenetic packages

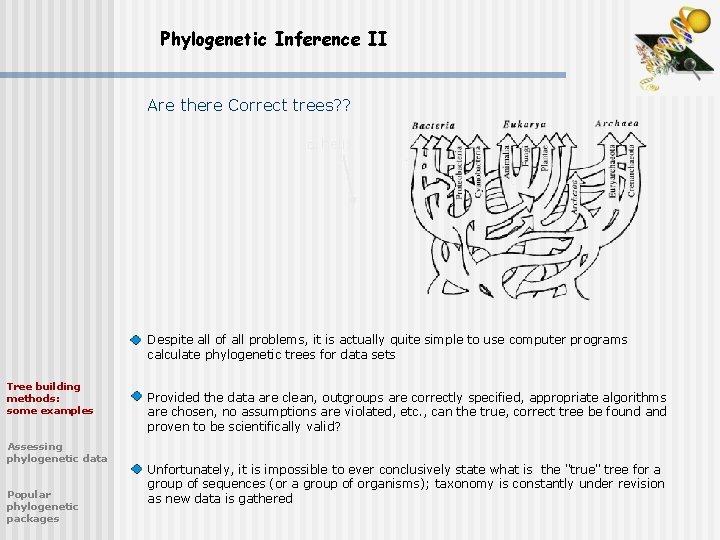
Phylogenetic Inference II Are there Correct trees? ? helix sheet Despite all of all problems, it is actually quite simple to use computer programs calculate phylogenetic trees for data sets Tree building methods: some examples Assessing phylogenetic data Popular phylogenetic packages Provided the data are clean, outgroups are correctly specified, appropriate algorithms are chosen, no assumptions are violated, etc. , can the true, correct tree be found and proven to be scientifically valid? Unfortunately, it is impossible to ever conclusively state what is the "true" tree for a group of sequences (or a group of organisms); taxonomy is constantly under revision as new data is gathered

Phenetics versus cladistics Phenetic methods construct trees (phenograms) by considering the current states of characters without regard to the evolutionary history that brought the species to their current phenotypes; phenograms are based on overall similarity Tree building methods: some examples Assessing phylogenetic data Popular phylogenetic packages Cladistic methods construct trees (cladograms) rely on assumptions about ancestral relationships as well as on current data; cladograms are based on character evolution (e. g. shared derived characters)

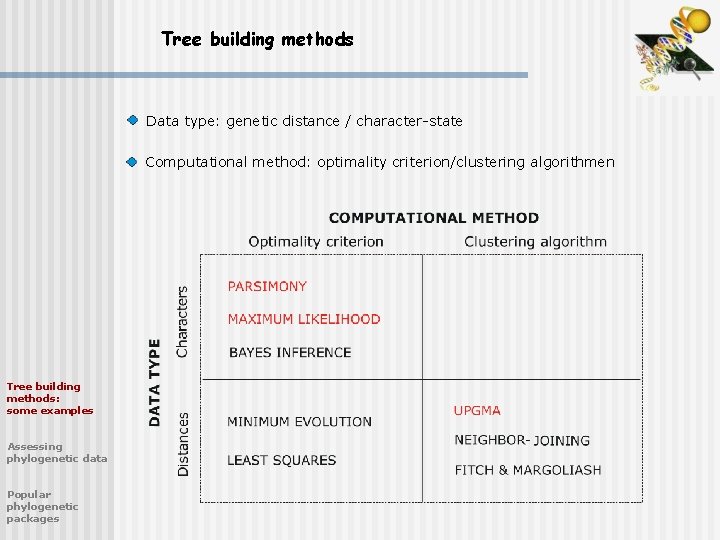
Tree building methods Data type: genetic distance / character-state Computational method: optimality criterion/clustering algorithmen Tree building methods: some examples Assessing phylogenetic data Popular phylogenetic packages

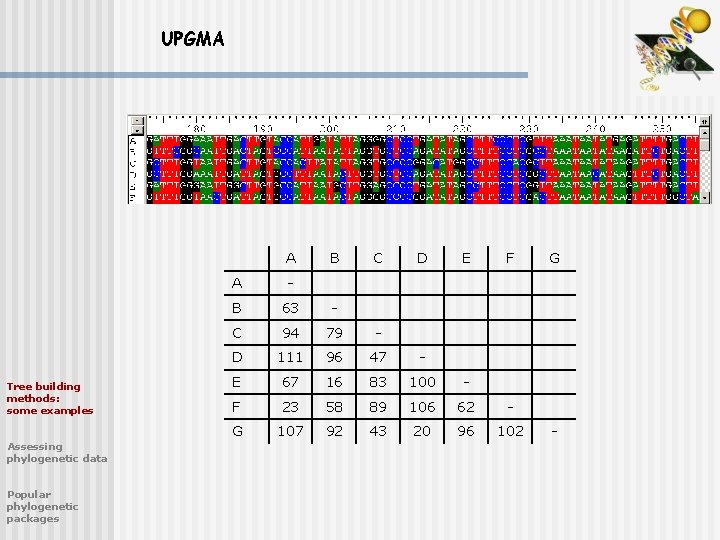
Tree building (distance based) UPGMA - The simplest of the distance methods is the UPGMA (Unweighted Pair Group Method using Arithmetic averages) - Many multiple alignment programs such as PILEUP use a variant of UPGMA to create a dendrogram of DNA sequences which is then used to guide the multiple alignment algorithm Tree building methods: some examples Assessing phylogenetic data Popular phylogenetic packages

UPGMA A Tree building methods: some examples Assessing phylogenetic data Popular phylogenetic packages B C D E F A - B 63 - C 94 79 - D 111 96 47 - E 67 16 83 100 - F 23 58 89 106 62 - G 107 92 43 20 96 102 G -

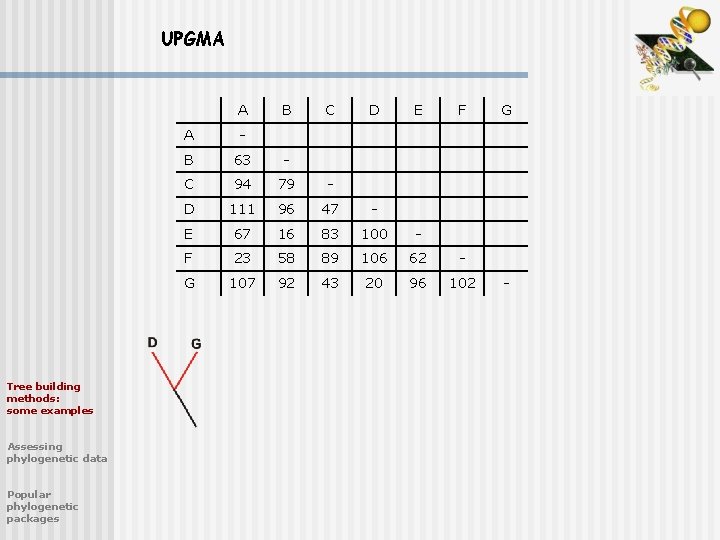
UPGMA A Tree building methods: some examples Assessing phylogenetic data Popular phylogenetic packages B C D E F A - B 63 - C 94 79 - D 111 96 47 - E 67 16 83 100 - F 23 58 89 106 62 - G 107 92 43 20 96 102 G -

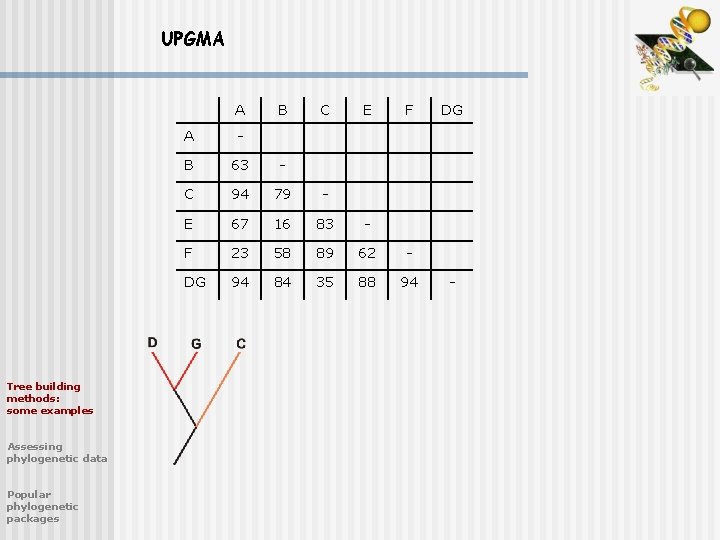
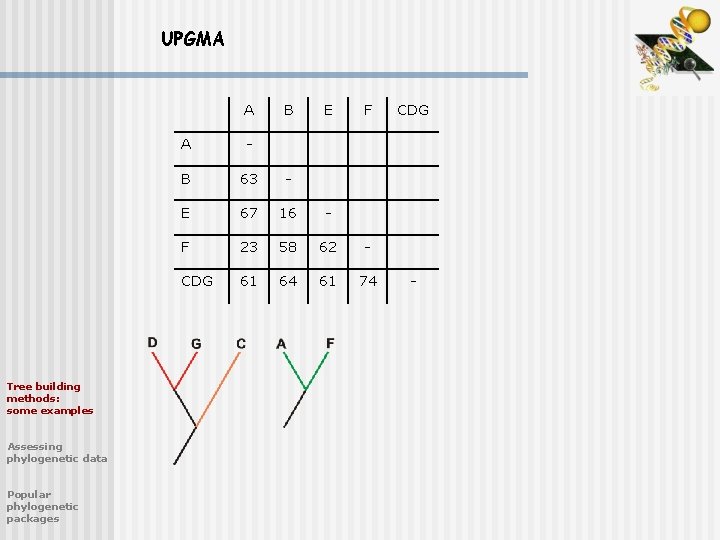
UPGMA A Tree building methods: some examples Assessing phylogenetic data Popular phylogenetic packages B C E F A - B 63 - C 94 79 - E 67 16 83 - F 23 58 89 62 - DG 94 84 35 88 94 DG -

UPGMA A Tree building methods: some examples Assessing phylogenetic data Popular phylogenetic packages B E F A - B 63 - E 67 16 - F 23 58 62 - CDG 61 64 61 74 CDG -

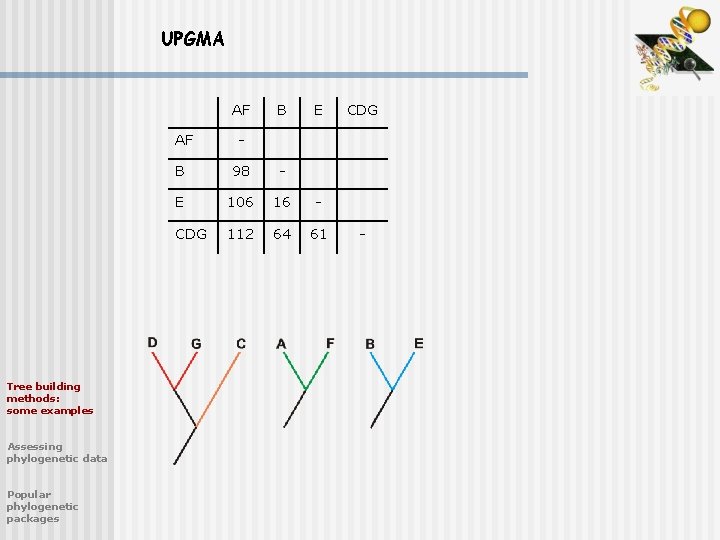
UPGMA AF Tree building methods: some examples Assessing phylogenetic data Popular phylogenetic packages B E AF - B 98 - E 106 16 - CDG 112 64 61 CDG -

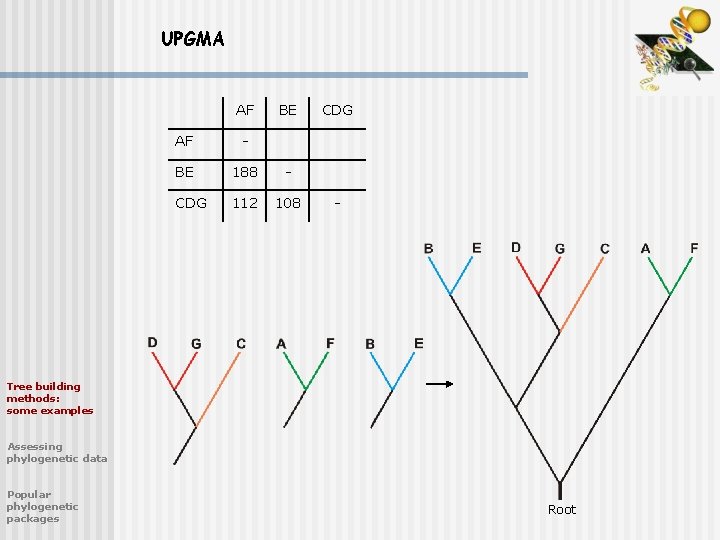
UPGMA AF BE AF - BE 188 - CDG 112 108 CDG - Tree building methods: some examples Assessing phylogenetic data Popular phylogenetic packages Root

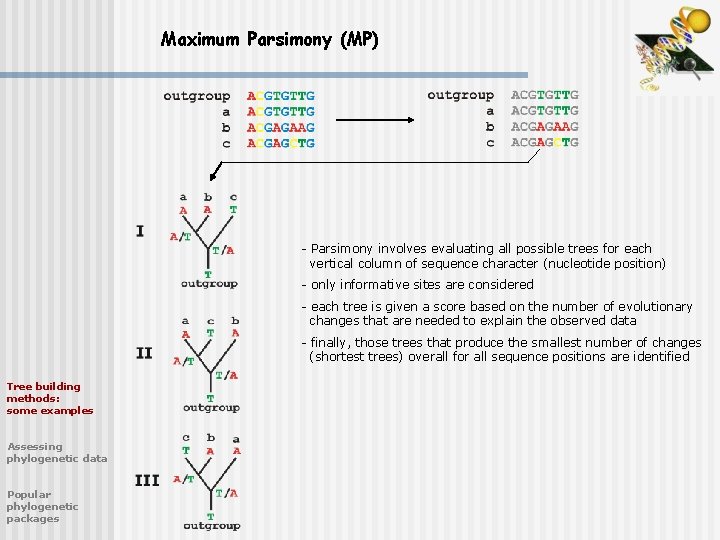
Maximum Parsimony (MP) - Parsimony involves evaluating all possible trees for each vertical column of sequence character (nucleotide position) - only informative sites are considered - each tree is given a score based on the number of evolutionary changes that are needed to explain the observed data - finally, those trees that produce the smallest number of changes (shortest trees) overall for all sequence positions are identified Tree building methods: some examples Assessing phylogenetic data Popular phylogenetic packages

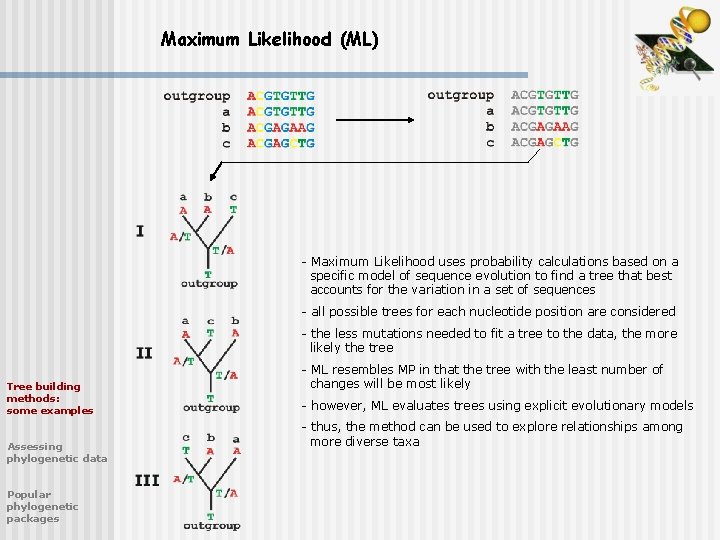
Maximum Likelihood (ML) - Maximum Likelihood uses probability calculations based on a specific model of sequence evolution to find a tree that best accounts for the variation in a set of sequences - all possible trees for each nucleotide position are considered - the less mutations needed to fit a tree to the data, the more likely the tree Tree building methods: some examples Assessing phylogenetic data Popular phylogenetic packages - ML resembles MP in that the tree with the least number of changes will be most likely - however, ML evaluates trees using explicit evolutionary models - thus, the method can be used to explore relationships among more diverse taxa

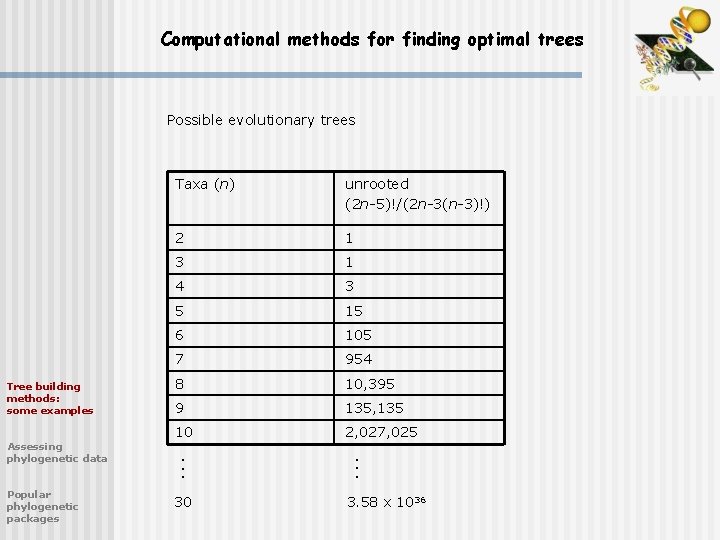
Computational methods for finding optimal trees Assessing phylogenetic data Popular phylogenetic packages unrooted (2 n-5)!/(2 n-3(n-3)!) 2 1 3 1 4 3 5 15 6 105 7 954 8 10, 395 9 135, 135 10 2, 027, 025. . . Tree building methods: some examples Taxa (n) . . . Possible evolutionary trees 30 3. 58 x 1036

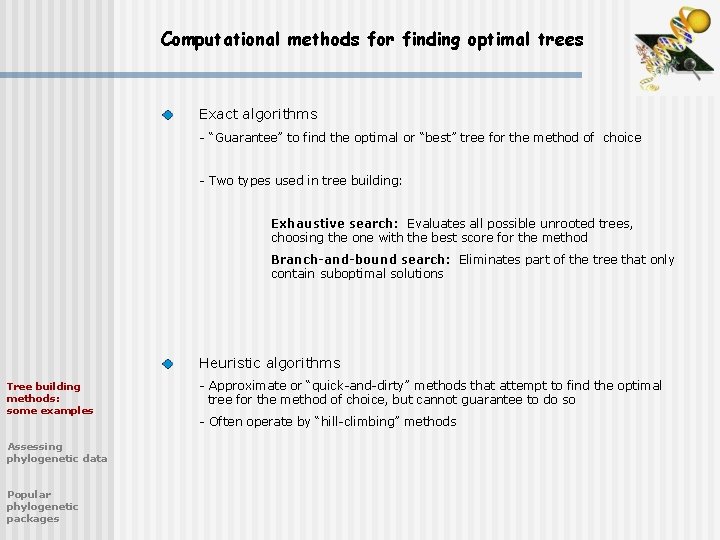
Computational methods for finding optimal trees Exact algorithms - “Guarantee” to find the optimal or “best” tree for the method of choice - Two types used in tree building: Exhaustive search: Evaluates all possible unrooted trees, choosing the one with the best score for the method Branch-and-bound search: Eliminates part of the tree that only contain suboptimal solutions Heuristic algorithms Tree building methods: some examples Assessing phylogenetic data Popular phylogenetic packages - Approximate or “quick-and-dirty” methods that attempt to find the optimal tree for the method of choice, but cannot guarantee to do so - Often operate by “hill-climbing” methods

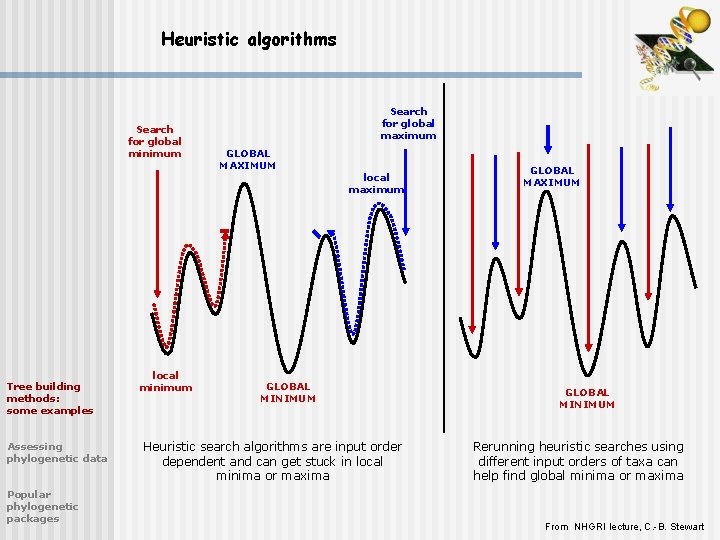
Heuristic algorithms Search for global minimum Tree building methods: some examples Assessing phylogenetic data Popular phylogenetic packages local minimum Search for global maximum GLOBAL MAXIMUM local maximum GLOBAL MINIMUM Heuristic search algorithms are input order dependent and can get stuck in local minima or maxima GLOBAL MAXIMUM GLOBAL MINIMUM Rerunning heuristic searches using different input orders of taxa can help find global minima or maxima From NHGRI lecture, C. -B. Stewart

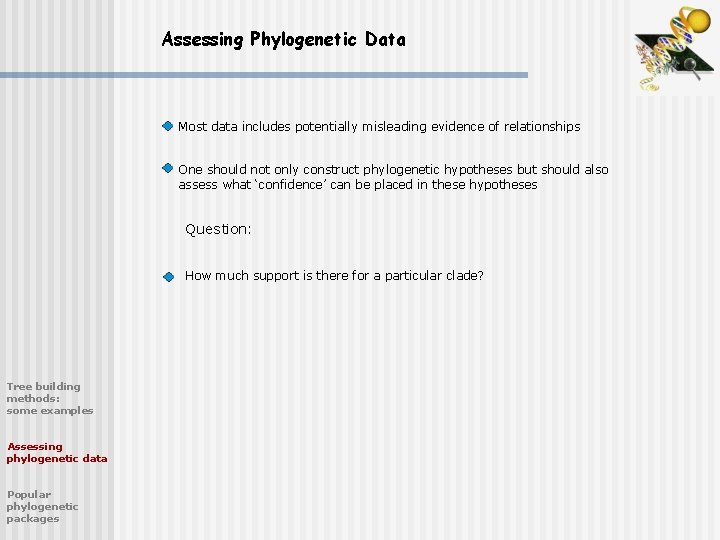
Assessing Phylogenetic Data Most data includes potentially misleading evidence of relationships One should not only construct phylogenetic hypotheses but should also assess what ‘confidence’ can be placed in these hypotheses Question: How much support is there for a particular clade? Tree building methods: some examples Assessing phylogenetic data Popular phylogenetic packages

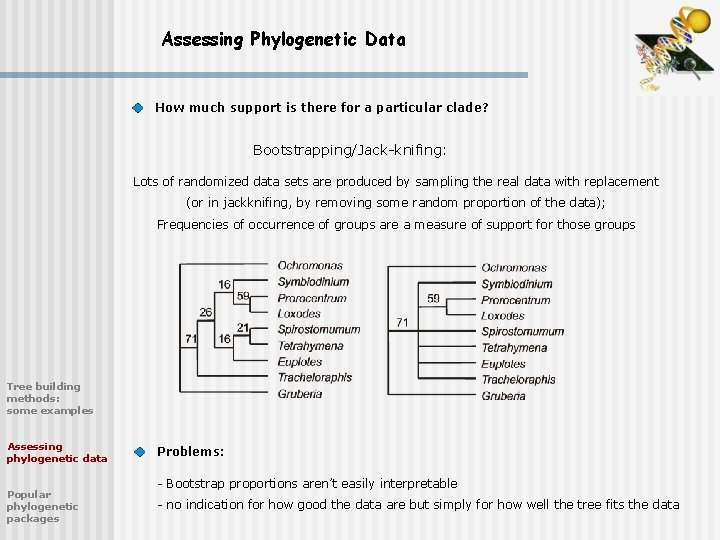
Assessing Phylogenetic Data How much support is there for a particular clade? Bootstrapping/Jack-knifing: Lots of randomized data sets are produced by sampling the real data with replacement (or in jackknifing, by removing some random proportion of the data); Frequencies of occurrence of groups are a measure of support for those groups Tree building methods: some examples Assessing phylogenetic data Popular phylogenetic packages Problems: - Bootstrap proportions aren’t easily interpretable - no indication for how good the data are but simply for how well the tree fits the data

Popular phylogenetic software packages Review available at: http: //evolution. genetics. washington. edu/phylip/software. html Tree building methods: some examples Assessing phylogenetic data Popular phylogenetic packages