Large scale DNA editing of retrotransposons accelerates mammalian

![An algorithm Define the transition probability: p=[#(C-to-T)+#(T-to-C)] / (2*alignment_length). k- cluster length, n- sequence An algorithm Define the transition probability: p=[#(C-to-T)+#(T-to-C)] / (2*alignment_length). k- cluster length, n- sequence](https://slidetodoc.com/presentation_image_h2/cec31c4de2325b9ef2cfd0aceb9a83bf/image-9.jpg)
![An algorithm Define the transition probability: p=[#(C-to-T)+#(T-to-C)] / (2*alignment_length). k- cluster length, n- sequence An algorithm Define the transition probability: p=[#(C-to-T)+#(T-to-C)] / (2*alignment_length). k- cluster length, n- sequence](https://slidetodoc.com/presentation_image_h2/cec31c4de2325b9ef2cfd0aceb9a83bf/image-10.jpg)







- Slides: 26

Large scale DNA editing of retrotransposons accelerates mammalian genome evolution Shai Carmi, Erez Levanon Bar-Ilan University 2010

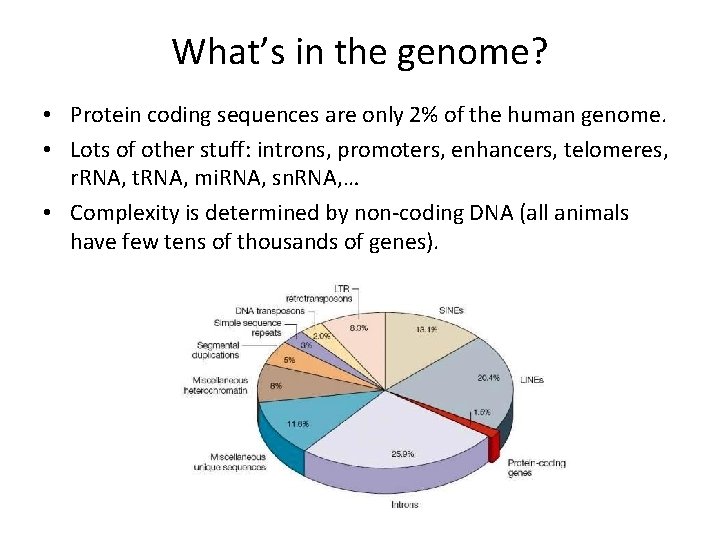
What’s in the genome? • Protein coding sequences are only 2% of the human genome. • Lots of other stuff: introns, promoters, enhancers, telomeres, r. RNA, t. RNA, mi. RNA, sn. RNA, … • Complexity is determined by non-coding DNA (all animals have few tens of thousands of genes).

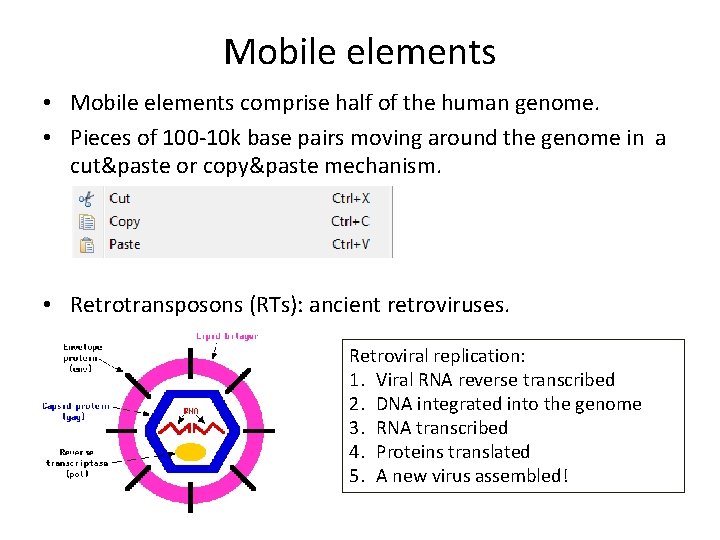
Mobile elements • Mobile elements comprise half of the human genome. • Pieces of 100 -10 k base pairs moving around the genome in a cut&paste or copy&paste mechanism. • Retrotransposons (RTs): ancient retroviruses. Retroviral replication: 1. Viral RNA reverse transcribed 2. DNA integrated into the genome 3. RNA transcribed 4. Proteins translated 5. A new virus assembled!

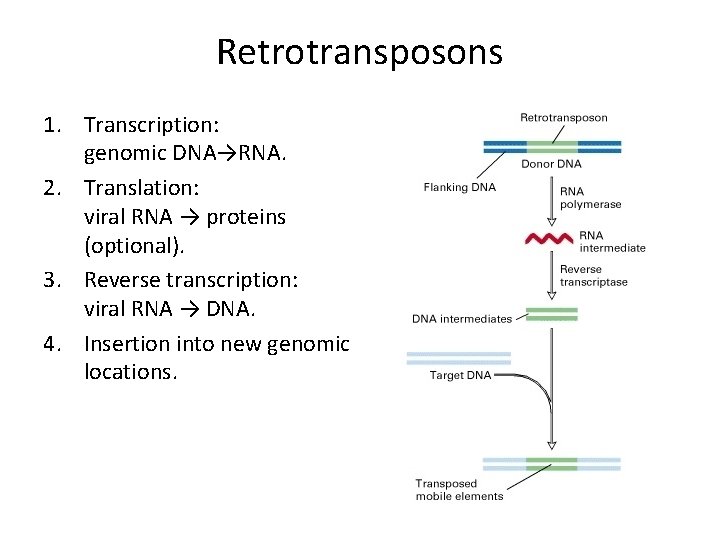
Retrotransposons 1. Transcription: genomic DNA→RNA. 2. Translation: viral RNA → proteins (optional). 3. Reverse transcription: viral RNA → DNA. 4. Insertion into new genomic locations.

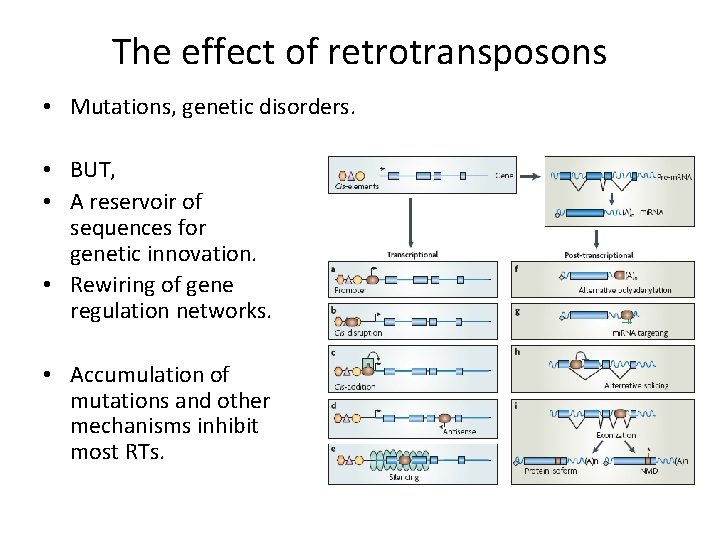
The effect of retrotransposons • Mutations, genetic disorders. • BUT, • A reservoir of sequences for genetic innovation. • Rewiring of gene regulation networks. • Accumulation of mutations and other mechanisms inhibit most RTs.

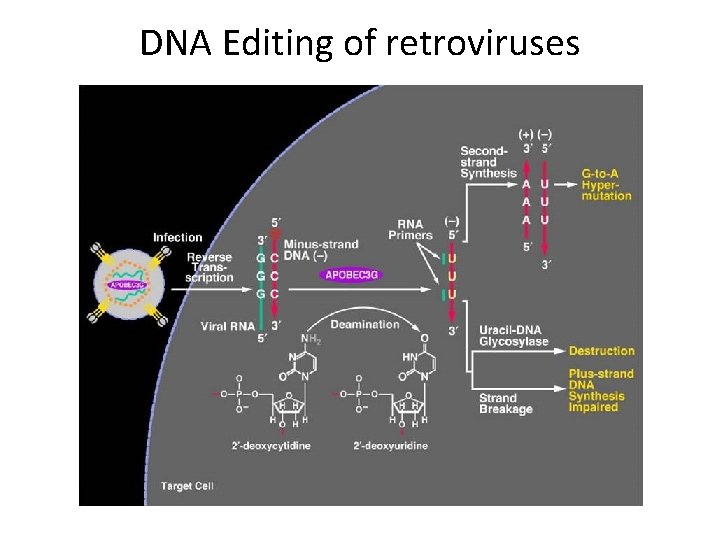
DNA Editing of retroviruses

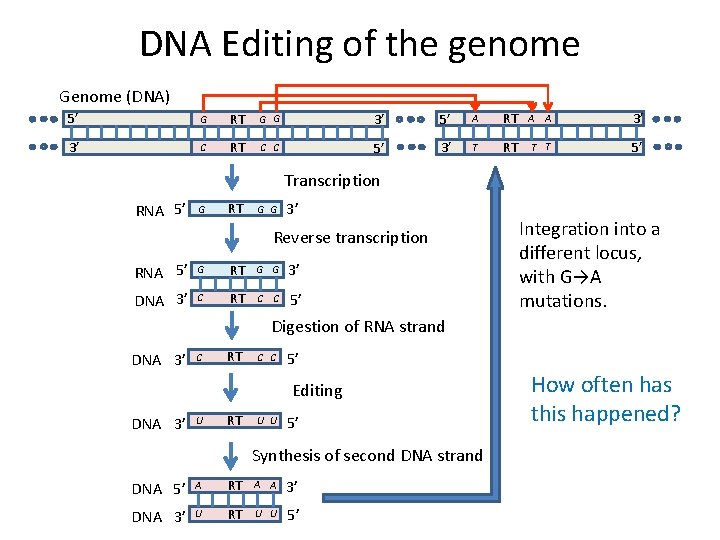
DNA Editing of the genome Genome (DNA) 5’ G RT G G 3’ 5’ A RT A A 3’ 3’ C RT C C 5’ 3’ T RT T T 5’ Transcription RNA 5’ G RT G G 3’ Reverse transcription RNA 5’ G RT G G 3’ DNA 3’ C RT C C 5’ Integration into a different locus, with G→A mutations. Digestion of RNA strand DNA 3’ C RT C C 5’ Editing DNA 3’ U RT U U 5’ Synthesis of second DNA strand DNA 5’ A RT A A 3’ DNA 3’ U RT U U 5’ How often has this happened?

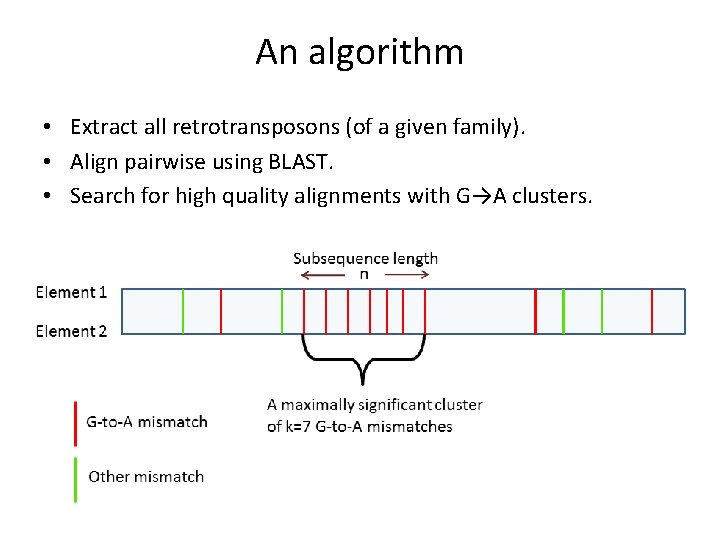
An algorithm • Extract all retrotransposons (of a given family). • Align pairwise using BLAST. • Search for high quality alignments with G→A clusters.
![An algorithm Define the transition probability pCtoTTtoC 2alignmentlength k cluster length n sequence An algorithm Define the transition probability: p=[#(C-to-T)+#(T-to-C)] / (2*alignment_length). k- cluster length, n- sequence](https://slidetodoc.com/presentation_image_h2/cec31c4de2325b9ef2cfd0aceb9a83bf/image-9.jpg)
An algorithm Define the transition probability: p=[#(C-to-T)+#(T-to-C)] / (2*alignment_length). k- cluster length, n- sequence length.
![An algorithm Define the transition probability pCtoTTtoC 2alignmentlength k cluster length n sequence An algorithm Define the transition probability: p=[#(C-to-T)+#(T-to-C)] / (2*alignment_length). k- cluster length, n- sequence](https://slidetodoc.com/presentation_image_h2/cec31c4de2325b9ef2cfd0aceb9a83bf/image-10.jpg)
An algorithm Define the transition probability: p=[#(C-to-T)+#(T-to-C)] / (2*alignment_length). k- cluster length, n- sequence length. • How many clusters do we expect by chance? • Use p=[#(G→A)+#(A→G)] / (2*alignment_length), and search for clusters of C→T! • Editing is strand-specific, and we align only positive strands. • True DNA editing will show no C→T clusters.

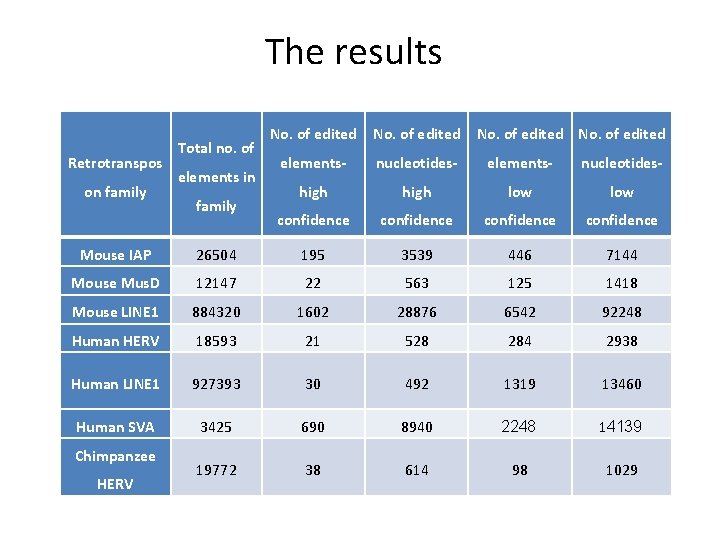
The results Retrotranspos on family Total no. of elements in family No. of edited elements- nucleotides- high low confidence Mouse IAP 26504 195 3539 446 7144 Mouse Mus. D 12147 22 563 125 1418 Mouse LINE 1 884320 1602 28876 6542 92248 Human HERV 18593 21 528 284 2938 Human LINE 1 927393 30 492 1319 13460 Human SVA 3425 690 8940 2248 14139 19772 38 614 98 1029 Chimpanzee HERV

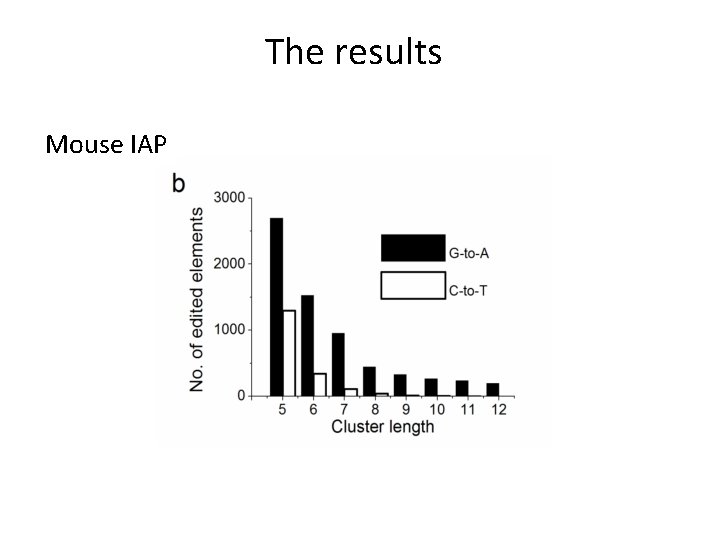
The results Mouse IAP

An example Mouse chr 8: 28575443 -28581824 (6, 382 nts) vs. chr 9: 114987516 -114993954. 176 G→A mismatches and only 26 other mismatches.

More examples Mouse IAP Query Sbjct Query Sbjct 4059 960 4119 1020 4179 1080 4239 1140 4299 1200 4359 1260 4419 1320 4479 1380 AAAACTGGCATAGGTGCCTATGTGGCTAATGGTAAAGTGGTATCCAAACAATATAATGAA. . . . A. . AATTCACCTCAAGTGGTAGAATGTTTAGTGGTCTTAGAAGTTTTAAAAACCTTTTTAAAA. . . . A. . . CCCCTTAATATTGTGTCAGATTCCTGTTATGTGGTTAATGCAGTAAATCTTTTAGAAGTG. . . A. . GCTGGAGTGATTAAGCCTTCCAGTAGAGTTGCCAATATTTTTCAGCAGATACAATTAGTT. . . A. . . . TTGTTATCTAGAAGATCTCCTGTTTATATTACTCATGTTAGAGCCCATTCAGGCCTACCT. . . . . A. . . . . GGCCCCATGGCTCTGGGAAATGATTTGGCAGATAAGGCCACTAAAGTGGTGGCTGCTGCC. . . AAA. . . . . CTATCATCCCCGGTAGAGGCTGCAAGAAATTTTCATAACAATTTTCATGTGACGGCTGAA. . . . A. . ACATTACGCAGTCGTTTCTCCTTGACAAGAAGCCCGTGACATTGTTACTCAATGT. . . A. . . Query Sbjct Query Sbjct 1381 1441 1501 1561 1621 1681 4118 1019 4178 1079 4238 1139 4298 1199 4358 1259 4418 1319 4478 1379 4538 1439 Mouse Mus. D GCCGCACGCCGTGCTTGGGGAAGGTTGCCTGTCAAAGGAGAGATTGGTGGAAGTTTAGCT. . . A. . . AA. . AGCATTCGGCAGAGTTCTGATGAACCATATCAGGATTTTGTGGACAGGCTATTGATTTCA. A. . . . GCTAGTAGAATCCTTGGAAATCCGGACACGGGAAGTCCTTTCGTTATGCAATTGGCTTAT. . . AA. . . . . GAGAATGCTAATGCAATTTGCCGAGCTGCGATTCAACCGCATAAGGGAACGACAGATTTG. . . A. . . GCGGGATATGTCCGCCTTTGCACAGACATCGGGCCTTCCTGCGAGACCTTGCAGGGAACC. . . . A. . CACGCGCAGGCAATGTTCTCAAGGAAACGAGGGAAAAATGTATGCTTTAAGTGTGGAAGT. . . . A. . . . . 1440 1500 1560 1620 1680 1740

More examples Human HERV Query Sbjct Query Sbjct 235 1256 294 1315 353 1375 413 1435 473 1495 532 1554 TCCTTTAAACAAGGAACAGGTTAGACAAGCCTTTATCAATTCTGGTGCATGGA-AGATTG. . . AAT. . -A. C. A. . ATCTTGCTGATTTTGT-GAGAATTATTGACAGTCATTACCCAAAAATCTTCCAG G. . A. A. . . . . TTTTAAAAATTGACTACTTGGATTTTACCTAAAAATGCCAGACATAAACCTTTAGAAAAT. . . . AA. . . T. A. . . GCTCTGACGGTATTTACTGATGGTTCCAGCAATGAAAAAGCAACTTACACCAGGCCAAAA A. . G. . . A. . . GAACGAGTCCTTGAAACTCAATGTCACTCGGCTCAAAGAGCAGAGTT-GTTGTTGTCAAT A. . . . . TAA. . . A. A. C. AC. . -. . T-CAGTGTTACAAAATTTTAATCAGCCTATTAACATTGTATCAGATTCTGCATATGTAGT. A. A. . . . . A. 293 1314 352 1374 412 1434 472 1494 531 1553 590 1613 Human SVA Query Sbjct Query Sbjct 300 412 360 472 420 532 480 592 540 652 600 712 TGCCGGGATTGCAGACGGAGTCTGGTTCGCTCGGTGGTGCCCAGGCTGGAGTG. . . . AA. . . . . CAGTGGCGTGGTCTCGGCTCGCTGCAGCCTCCATCTCCCGGCCGCCTTGGCCGCCC. . . . . A. . . . T. . . AGAGTGCCGAGATTGCAGCCTCTGCCCGGCCTCCACCCCGTCTGGGAGGTGGGGAGCGTC. A. . . . A. . AA. . . . TCTGCCTGGCCGCCCATCGTCTGGGACGTGGGGAGCCCCTCTGCCTGGCTGCCCAGTCTG. . T. . . . . A. . . . GAGGGTGGGGAGCATCTCTGCCCGGCCGCCATCCCGTCTGGGAGGTGGGGAGCGCCTCTT. . AA. . G. . . . . A. . . . CCCGGCAGCCATCTGGGAGGTGGGGAGCGTCTCTGCCCGGCCGCCCATCGTCTGA. . . . 359 471 419 531 479 591 539 651 599 711 659 771

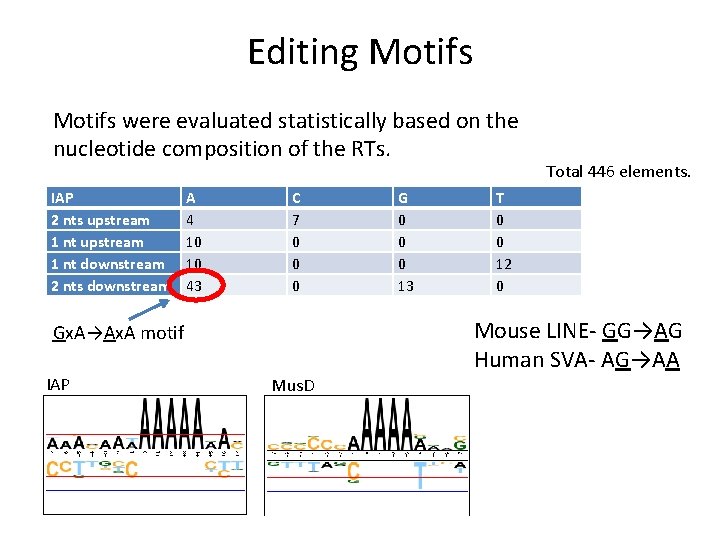
Editing Motifs were evaluated statistically based on the nucleotide composition of the RTs. IAP 2 nts upstream 1 nt downstream 2 nts downstream A 4 10 10 43 C 7 0 0 0 T 0 0 12 0 Mouse LINE- GG→AG Human SVA- AG→AA Gx. A→Ax. A motif IAP G 0 0 0 13 Total 446 elements. Mus. D

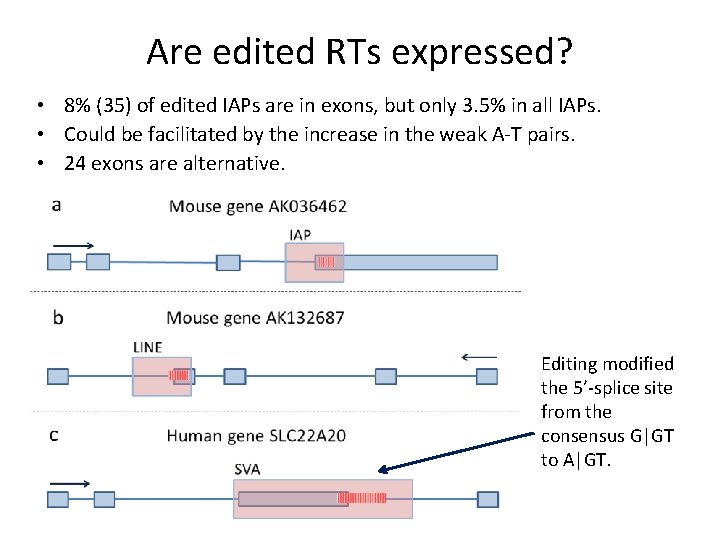
Are edited RTs expressed? • 8% (35) of edited IAPs are in exons, but only 3. 5% in all IAPs. • Could be facilitated by the increase in the weak A-T pairs. • 24 exons are alternative. Editing modified the 5’-splice site from the consensus G|GT to A|GT.

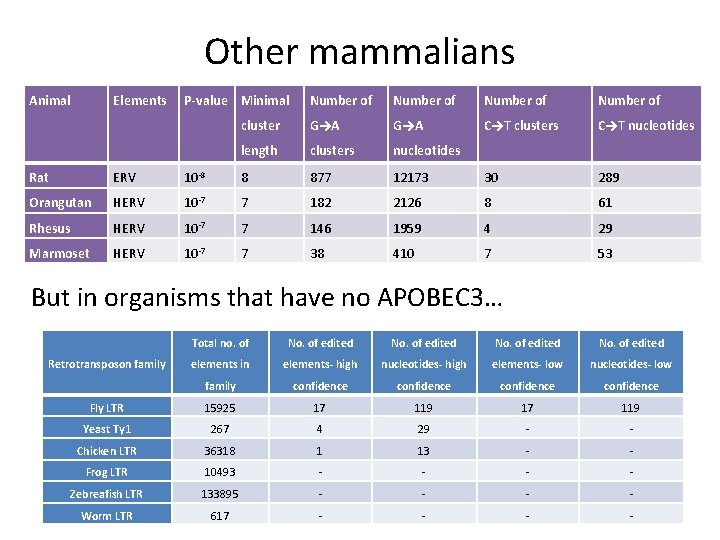
Other mammalians Animal Elements P-value Minimal Number of cluster G→A C→T clusters C→T nucleotides length clusters nucleotides Rat ERV 10 -8 8 877 12173 30 289 Orangutan HERV 10 -7 7 182 2126 8 61 Rhesus HERV 10 -7 7 146 1959 4 29 Marmoset HERV 10 -7 7 38 410 7 53 But in organisms that have no APOBEC 3… Total no. of No. of edited elements in elements- high nucleotides- high elements- low nucleotides- low family confidence Fly LTR 15925 17 119 Yeast Ty 1 267 4 29 - - Chicken LTR 36318 1 13 - - Frog LTR 10493 - - Zebreafish LTR 133895 - - Worm LTR 617 - - Retrotransposon family

Editing is ongoing • • SVA RTs are hominoid-specific Largest fraction of elements are edited (690, 20%) 262 human-specific edited elements 16 polymorphic elements

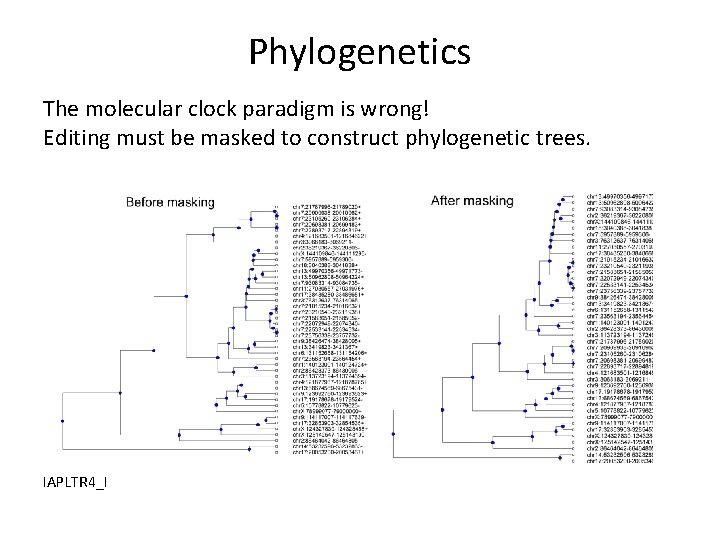
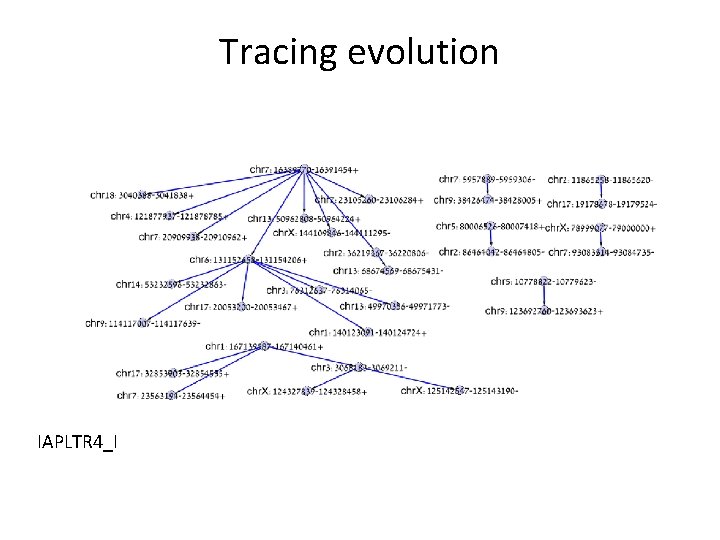
Phylogenetics The molecular clock paradigm is wrong! Editing must be masked to construct phylogenetic trees. IAPLTR 4_I

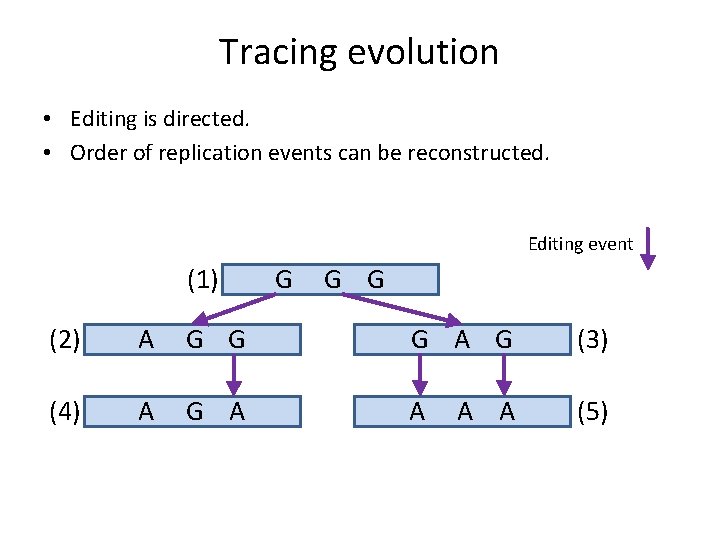
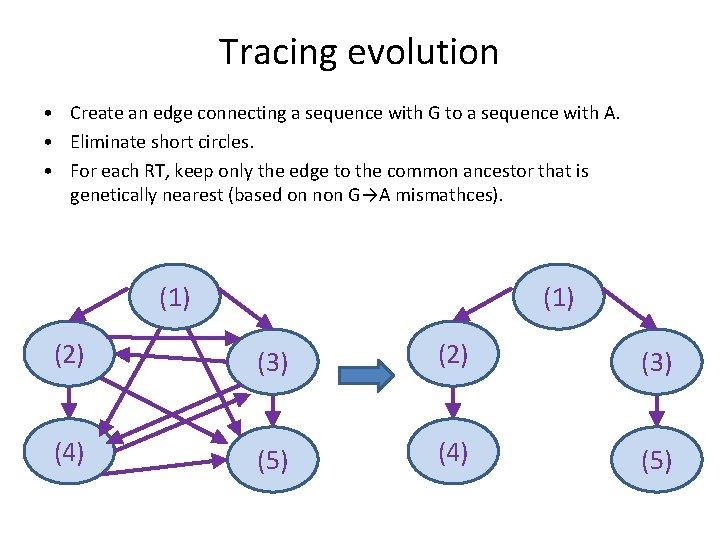
Tracing evolution • Editing is directed. • Order of replication events can be reconstructed. Editing event (1) G G G (2) A G G G A G (3) (4) A G A A (5) A A

Tracing evolution • Create an edge connecting a sequence with G to a sequence with A. • Eliminate short circles. • For each RT, keep only the edge to the common ancestor that is genetically nearest (based on non G→A mismathces). (1) (2) (3) (4) (5)

Tracing evolution IAPLTR 4_I

Discussion • • Editing can explain the successful exaptation of RTs Editing accelerates evolution- demonstrated for HIV Our method detects only a small fraction of edited elements De novo genes from edited RTs probably not here yet

Future directions • An editing-based algorithm to reconstruct the history of retrotransposon evolution. • A comprehensive survey of editing in the reference genome. • A systematic search for functions of edited elements (expression with RNA-seq, positive selection). • Searching for editing in non-reference DNA: o Different individuals (polymorphism). o Different tissues (somatic editing).

Thank you CGACAAGAGTGTACGATGACGTC |||||*|||||*|||| CGACCGGAGTGTGCGCTGGCGTC
 Central editing in research methodology
Central editing in research methodology Nonlinear video editing
Nonlinear video editing What is a line scale
What is a line scale Large scale vs small scale map
Large scale vs small scale map Map scales definition
Map scales definition Map scale ratio
Map scale ratio Physical and cultural features of topographic map
Physical and cultural features of topographic map Maintenance of body temperature
Maintenance of body temperature Excretory organ of mammals
Excretory organ of mammals Mammalian excretory system
Mammalian excretory system Mammalian lung diagram
Mammalian lung diagram Tricodonta
Tricodonta Mammalian excretory system
Mammalian excretory system Mammalian biomanufacturing
Mammalian biomanufacturing Mammalian dive reflex
Mammalian dive reflex Chapter 30 section 1 mammalian characteristics answer key
Chapter 30 section 1 mammalian characteristics answer key Cartilage in trachea function
Cartilage in trachea function Tendinous cords
Tendinous cords Mammalian excretory system
Mammalian excretory system Describe how a baseball player accelerates
Describe how a baseball player accelerates Find the average acceleration of a northbound subway
Find the average acceleration of a northbound subway Newton dynamics
Newton dynamics Describing and measuring motion worksheet
Describing and measuring motion worksheet A car of mass 1000 kg accelerates
A car of mass 1000 kg accelerates Turner's treadmill runs with a velocity of -1.2 m/s
Turner's treadmill runs with a velocity of -1.2 m/s A rocket of mass 12000 kg accelerates vertically
A rocket of mass 12000 kg accelerates vertically Chapter 11 motion section 2 acceleration answer key
Chapter 11 motion section 2 acceleration answer key