June 10 th 2006 ETH Zurich Switzerland Organic







- Slides: 21

June 10 th, 2006 ETH Zurich, Switzerland Organic Molecules on Insulating Surfaces Investigated by NC-AFM Enrico Gnecco NCCR Nanoscale Science University of Basel, Switzerland

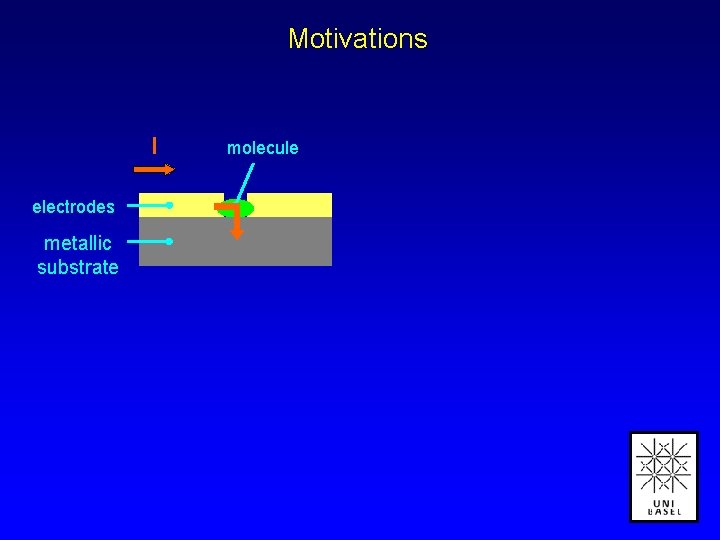
Motivations I electrodes metallic substrate molecule

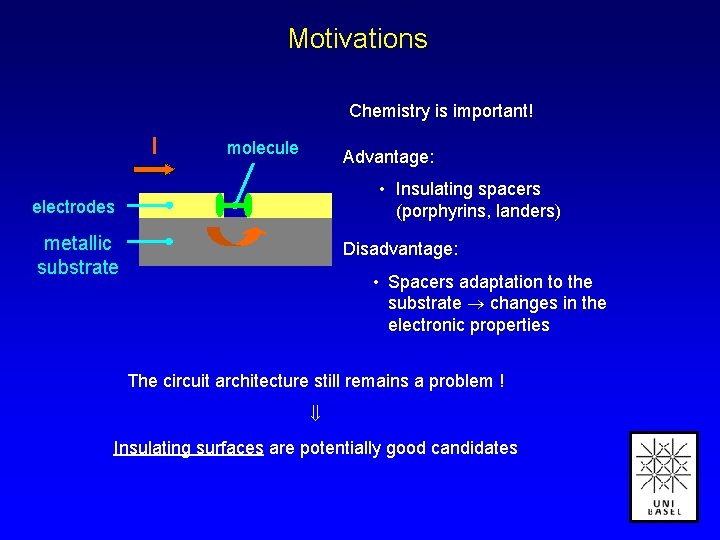
Motivations Chemistry is important! I molecule Advantage: • Insulating spacers (porphyrins, landers) electrodes metallic substrate Disadvantage: • Spacers adaptation to the substrate changes in the electronic properties The circuit architecture still remains a problem ! Insulating surfaces are potentially good candidates

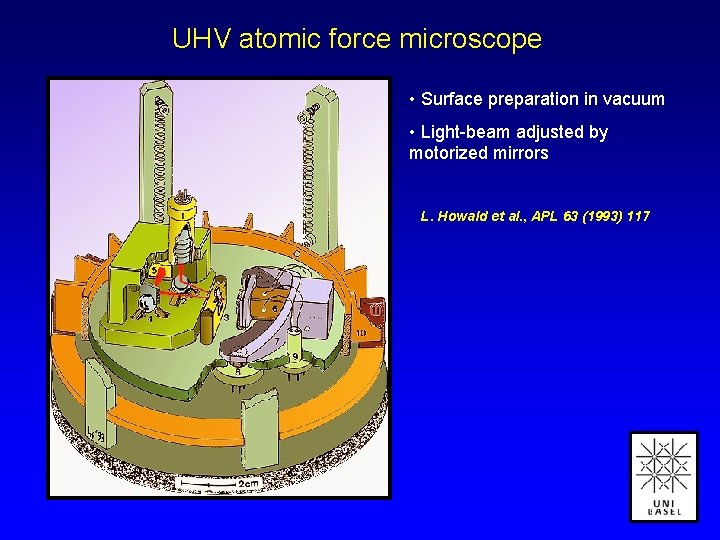
UHV atomic force microscope • Surface preparation in vacuum • Light-beam adjusted by motorized mirrors L. Howald et al. , APL 63 (1993) 117

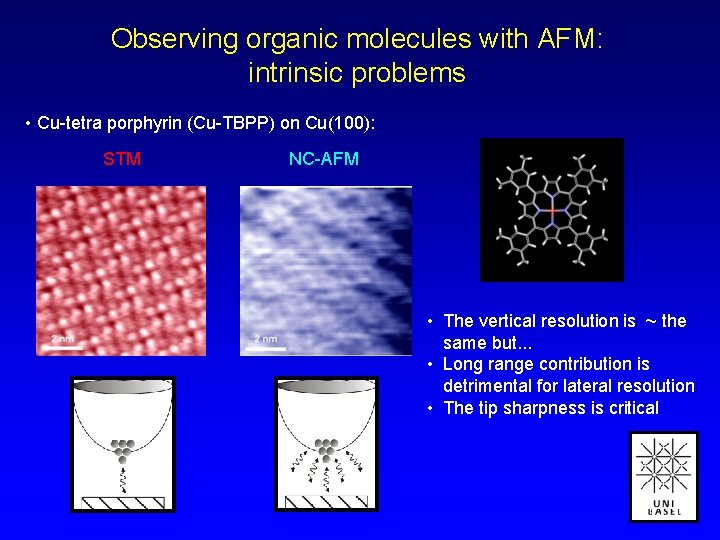
Observing organic molecules with AFM: intrinsic problems • Cu-tetra porphyrin (Cu-TBPP) on Cu(100): STM NC-AFM • The vertical resolution is ~ the same but. . . • Long range contribution is detrimental for lateral resolution • The tip sharpness is critical

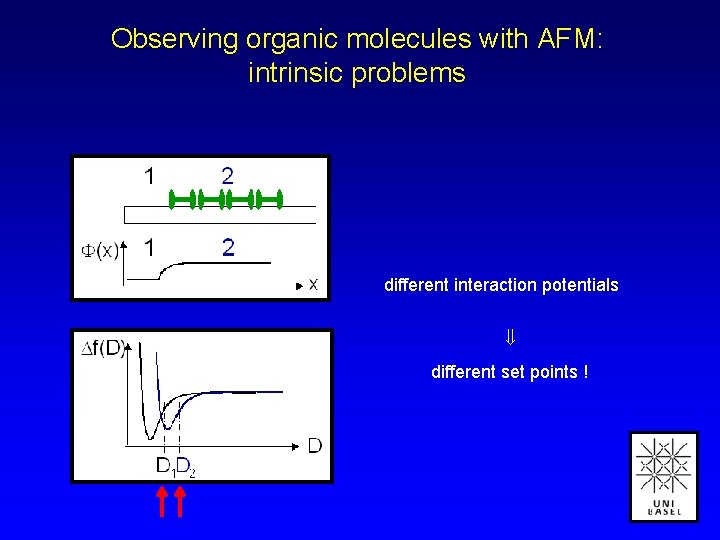
Observing organic molecules with AFM: intrinsic problems different interaction potentials different set points !

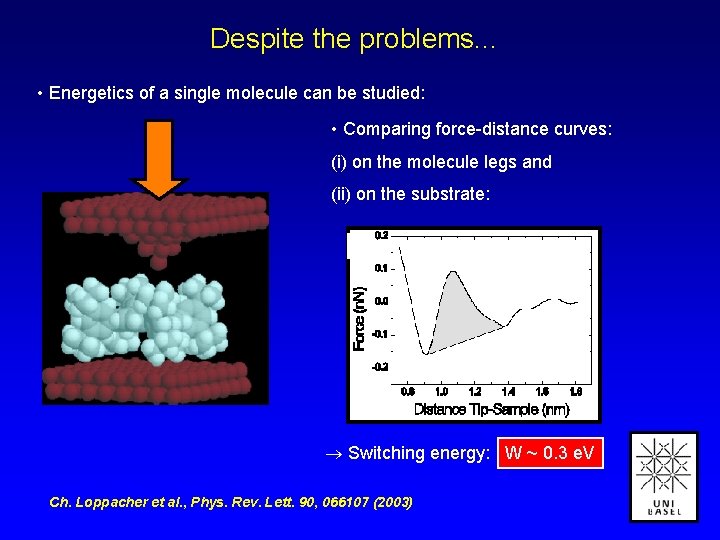
Despite the problems. . . • Energetics of a single molecule can be studied: • Comparing force-distance curves: (i) on the molecule legs and (ii) on the substrate: Switching energy: W ~ 0. 3 e. V Ch. Loppacher et al. , Phys. Rev. Lett. 90, 066107 (2003)

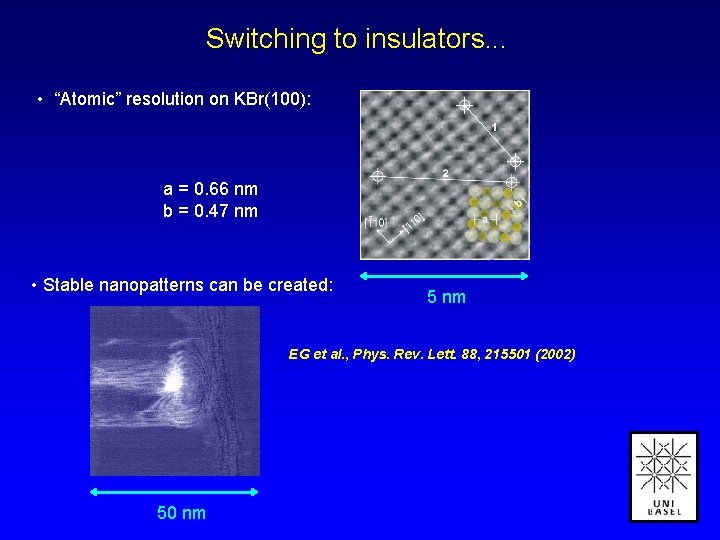
Switching to insulators. . . • “Atomic” resolution on KBr(100): a = 0. 66 nm b = 0. 47 nm • Stable nanopatterns can be created: 5 nm EG et al. , Phys. Rev. Lett. 88, 215501 (2002) 50 nm

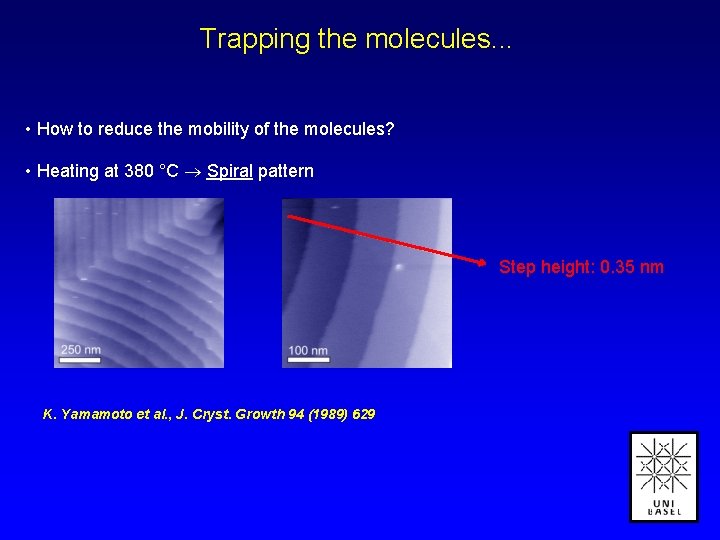
Trapping the molecules. . . • How to reduce the mobility of the molecules? • Heating at 380 °C Spiral pattern Step height: 0. 35 nm K. Yamamoto et al. , J. Cryst. Growth 94 (1989) 629

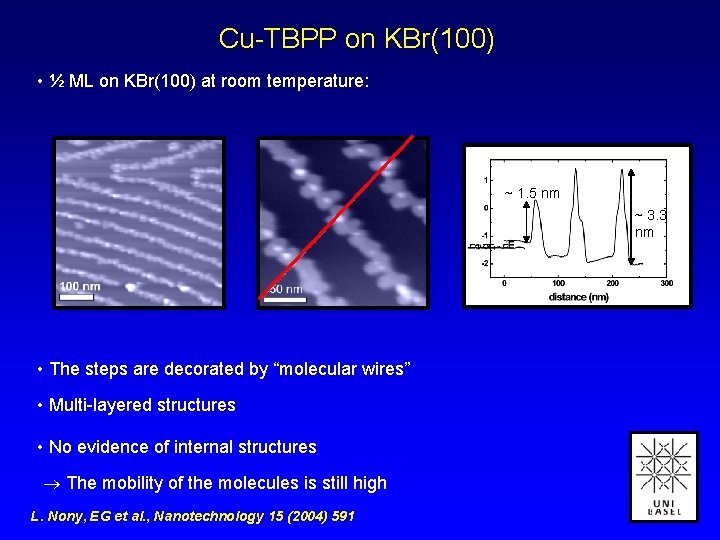
Cu-TBPP on KBr(100) • ½ ML on KBr(100) at room temperature: ~ 1. 5 nm ~ 3. 3 nm • The steps are decorated by “molecular wires” • Multi-layered structures • No evidence of internal structures The mobility of the molecules is still high L. Nony, EG et al. , Nanotechnology 15 (2004) 591

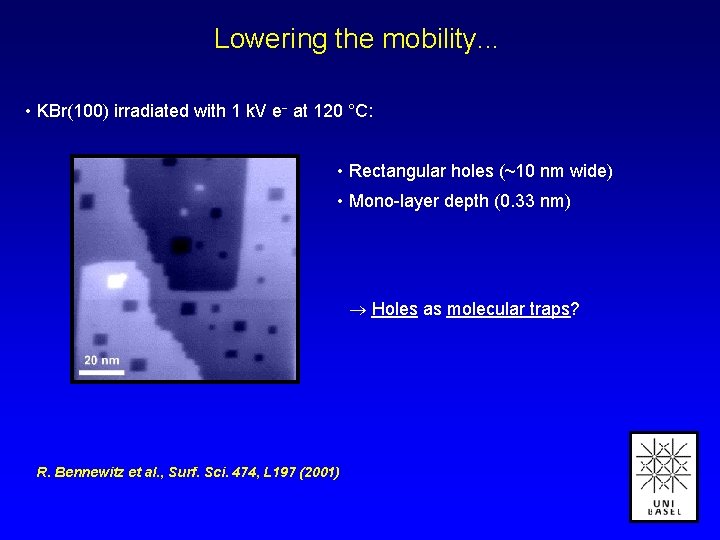
Lowering the mobility. . . • KBr(100) irradiated with 1 k. V e at 120 °C: • Rectangular holes (~10 nm wide) • Mono-layer depth (0. 33 nm) Holes as molecular traps? R. Bennewitz et al. , Surf. Sci. 474, L 197 (2001)

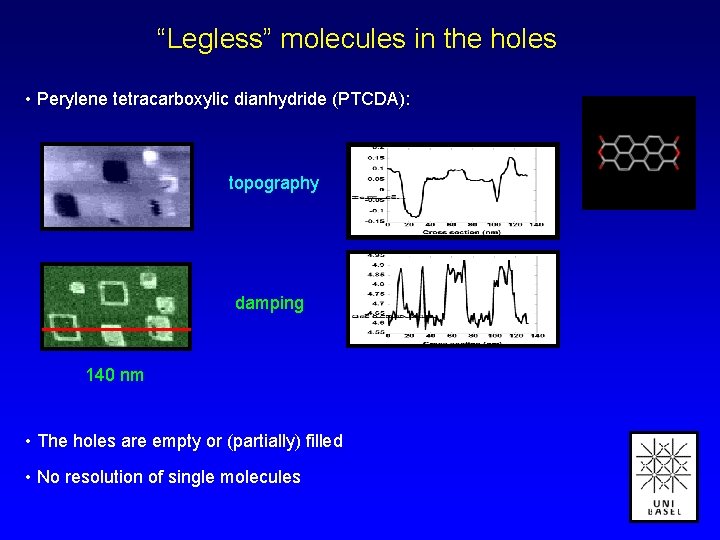
“Legless” molecules in the holes • Perylene tetracarboxylic dianhydride (PTCDA): topography damping 140 nm • The holes are empty or (partially) filled • No resolution of single molecules

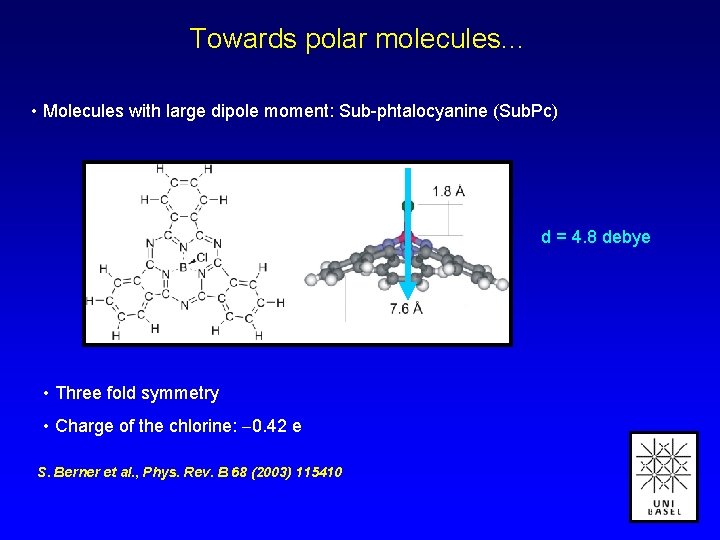
Towards polar molecules. . . • Molecules with large dipole moment: Sub-phtalocyanine (Sub. Pc) d = 4. 8 debye • Three fold symmetry • Charge of the chlorine: 0. 42 e S. Berner et al. , Phys. Rev. B 68 (2003) 115410

Sub. PC molecules on e -irradiated KBr • 1 ML on KBr(100) at 80 °C: 18 nm • Single molecules are resolved ! L. Nony, EG et al. , Nano Lett. 4 (2004) 2185

Molecular confinement • Some details: 1. 4 nm • The molecules are aligned in rows oriented 45° • Along some edges the molecules are mismatched • Height of the islands ~ 0. 6 nm (+ hole depth = 1 nm)

Matching the substrate. . . Potential arrangement of the molecules : • Apparent size ~1 nm • Alignment along the [110] axis • Regular rows: 3 b ~ 1. 4 nm • Distance between molecules in a row: 2 b ~ 0. 95 nm

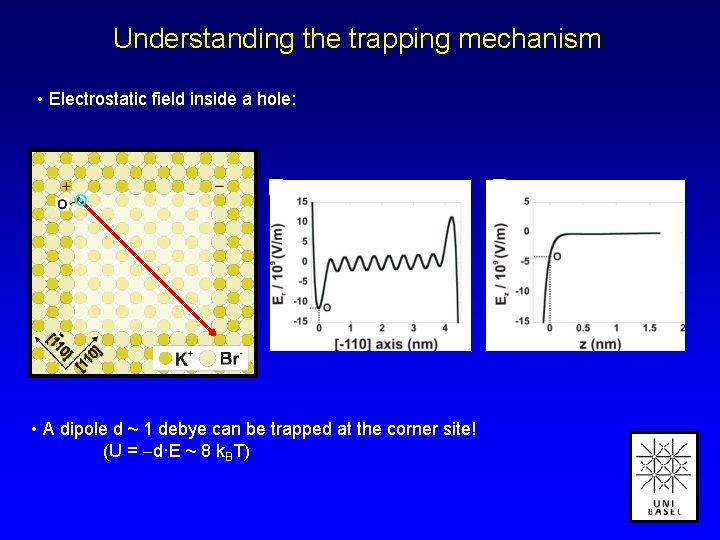
Understanding the trapping mechanism • Electrostatic field inside a hole: • A dipole d ~ 1 debye can be trapped at the corner site! (U = d·E ~ 8 k. BT)

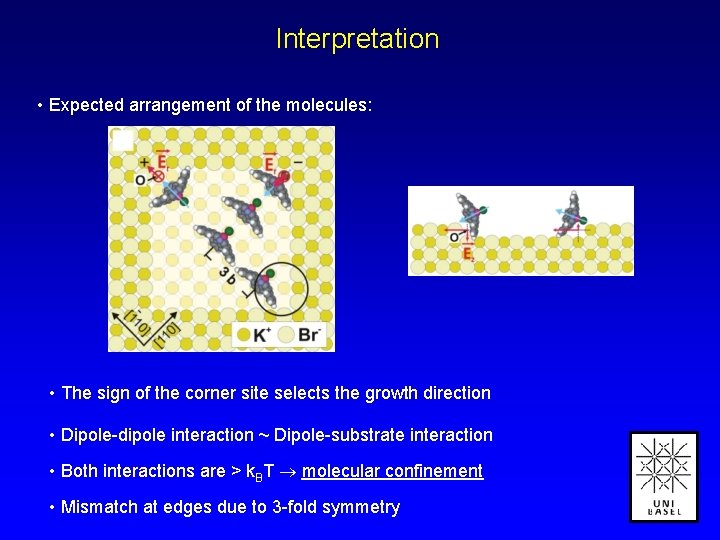
Interpretation • Expected arrangement of the molecules: • The sign of the corner site selects the growth direction • Dipole-dipole interaction ~ Dipole-substrate interaction • Both interactions are > k. BT molecular confinement • Mismatch at edges due to 3 -fold symmetry

Empty vs filled holes • On larger scale. . . 150 nm • Only the holes < 15 nm in size are filled !

Conclusions • Holes created by e irradiation on KBr act as molecular traps • Single organic molecules on insulators have been resolved by AFM • The size of the holes is critical Outlook • Molecules with 4 -fold symmetry • How to contact electrodes? • Theory of molecular confinement?

Acknowledgments UNI Basel University of Tokyo Ernst Meyer T. Eguchi Christoph Gerber Laurent Nony (*) CNRS Toulouse Alexis Baratoff A. Gourdon Roland Bennewitz (**) C. Joachim Oliver Pfeiffer Thomas Young (*) Now at Univ. Aix-Marseille III, France (**) Now at Mc. Gill Univ. , Montreal, Canada This work was supported by • The Swiss National Science Fundation • The Swiss National Center of Competence in Research on Nanoscale Science
 Eth meth
Eth meth Ethyl methyl chart
Ethyl methyl chart Wta formular zürich
Wta formular zürich Zoo zürich
Zoo zürich Phzh ilias
Phzh ilias Direktion der justiz und des innern kanton zürich
Direktion der justiz und des innern kanton zürich Universidad de zúrich egresados notables
Universidad de zúrich egresados notables Zurich
Zurich Einstein zurich notebook
Einstein zurich notebook Zwingli
Zwingli Gesuch installationsattest kanton zürich
Gesuch installationsattest kanton zürich Kinderurologie zürich
Kinderurologie zürich Kurs detailhandelsspezialistin
Kurs detailhandelsspezialistin Bfs winterthur
Bfs winterthur Ebi zürich
Ebi zürich Career crus
Career crus Zurich public sector
Zurich public sector Publikationsarten
Publikationsarten Zurich constance
Zurich constance Kindergarten gossau zh
Kindergarten gossau zh Matthias weisenhorn
Matthias weisenhorn Zurich subguard
Zurich subguard