Introduction to RNASeq and Transcriptome Analysis Hands on
















- Slides: 55

Introduction to RNA-Seq and Transcriptome Analysis Hands – on activities (Fun with UNIX!) Power. Point: Jessica Kirkpatrick and Casey Hanson RNA-Seq Lab | Jessica Kirkpatrick | 2015 1

Exercise 1. Use the Tuxedo Suite to: a. Align RNA-Seq reads using Top. Hat (splice-aware aligner). b. Perform reference-based transcriptome assembly with Cufflinks. c. Obtain a new transcriptome using Cufflinks & Cuffmerge. d. Use Cuffdiff to obtain a list of differentially expressed genes. e. Report a list of significantly expressed genes. 2. Use a genome browser and visualization tool to observe the aligned data and the new transcriptome. 2

Tuxedo Suite Bowtie and Bowtie use Burrows-Wheeler indexing for aligning reads. With bowtie 2 there is no upper limit on the read length Tophat uses either Bowtie or Bowtie 2 to align reads in a splice-aware manner and aids the discovery of new splice junctions The Cufflinks package has 4 components, the 2 major ones are listed below Cufflinks does reference-based transcriptome assembly Cuffdiff does statistical analysis and identifies differentially expressed transcripts in a simple pairwise comparison, and a series of pairwise comparisons in a time-course experiment Trapnell et al. , Nature Protocols, March 2012

Premise Question: Is there a difference in the results if the Tuxedo Suite is run 2 different ways? 1. Procedure: Run 1: Allow Top. Hat to select splice junctions and proceed through the steps without giving the software any information about known genes/gene models. Run 2: Force Top. Hat to use only known splice junctions (i. e. known genes/gene models) and proceed through the steps making sure we are doing our analysis in the context of these gene models. 2. Evaluation: a. 2 metrics: # of mapped reads and # of significantly different identified genes b. Compare new transcriptome to known genes. 4

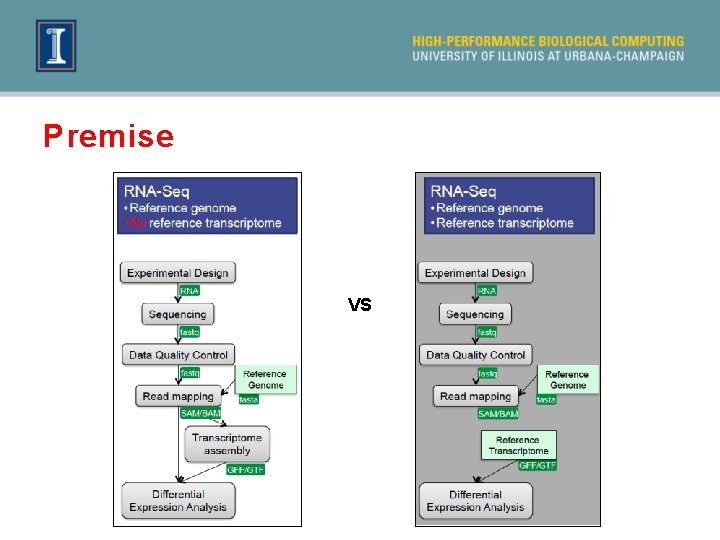
Premise VS

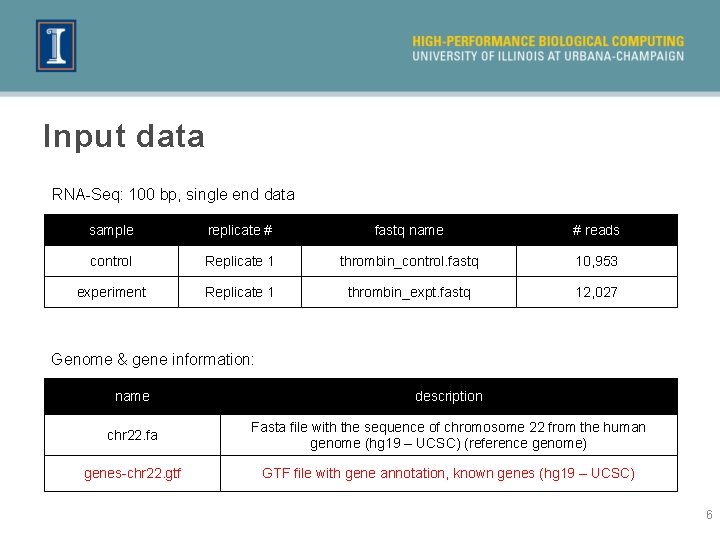
Input data RNA-Seq: 100 bp, single end data sample replicate # fastq name # reads control Replicate 1 thrombin_control. fastq 10, 953 experiment Replicate 1 thrombin_expt. fastq 12, 027 Genome & gene information: name description chr 22. fa Fasta file with the sequence of chromosome 22 from the human genome (hg 19 – UCSC) (reference genome) genes-chr 22. gtf GTF file with gene annotation, known genes (hg 19 – UCSC) 6

Sign in to Galaxy Go to https: //galaxy. illinois. edu Click on the button Sign in using your classroom ID and password

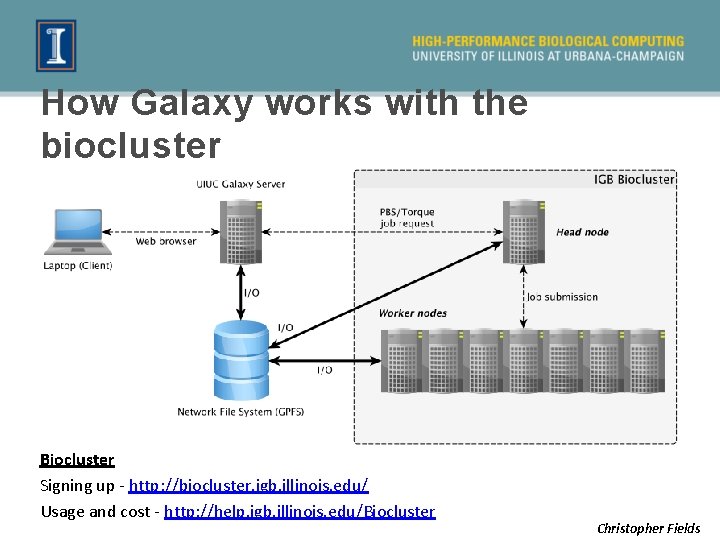
How Galaxy works with the biocluster Biocluster Signing up - http: //biocluster. igb. illinois. edu/ Usage and cost - http: //help. igb. illinois. edu/Biocluster Christopher Fields

Rename the History

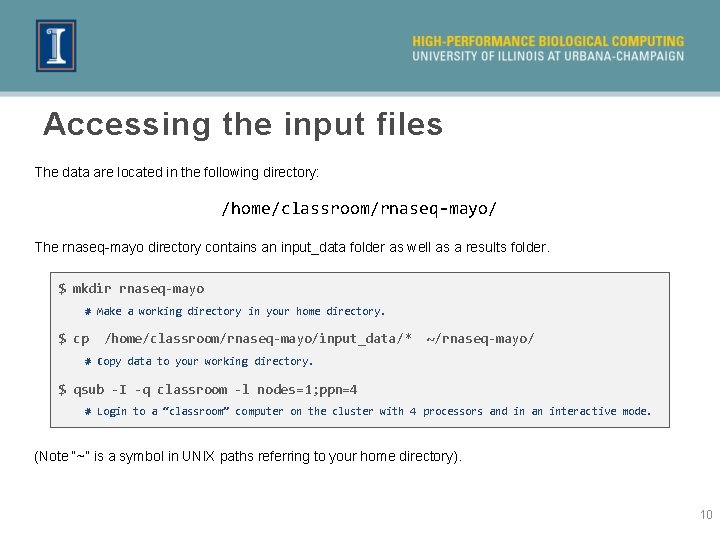
Accessing the input files The data are located in the following directory: /home/classroom/rnaseq-mayo/ The rnaseq-mayo directory contains an input_data folder as well as a results folder. $ mkdir rnaseq-mayo # Make a working directory in your home directory. $ cp /home/classroom/rnaseq-mayo/input_data/* ~/rnaseq-mayo/ # Copy data to your working directory. $ qsub -I -q classroom -l nodes=1; ppn=4 # Login to a “classroom” computer on the cluster with 4 processors and in an interactive mode. (Note “~” is a symbol in UNIX paths referring to your home directory). 10

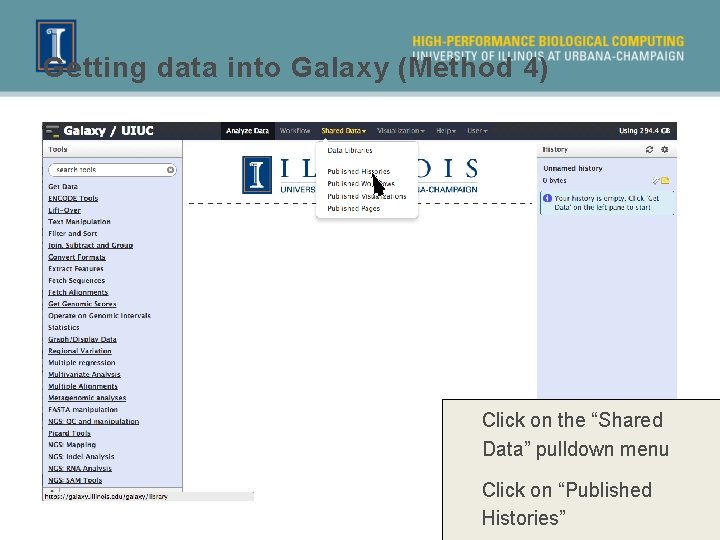
Getting data into Galaxy (Method 4) Click on the “Shared Data” pulldown menu Click on “Published Histories”

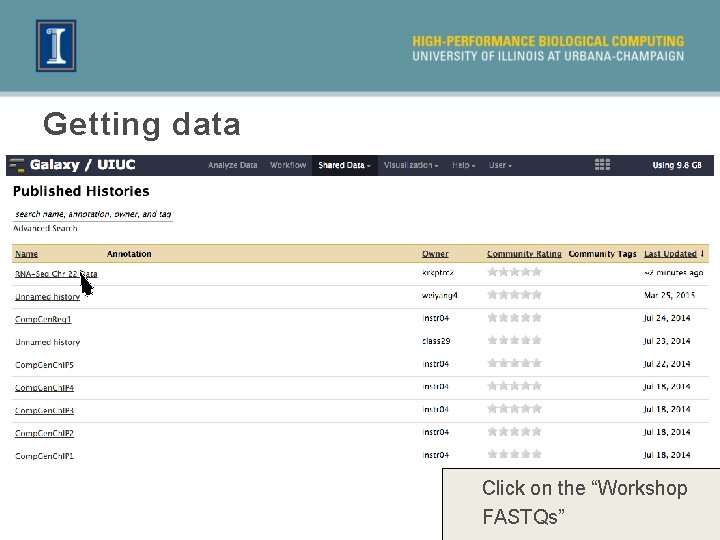
Getting data Click on the “Workshop FASTQs”

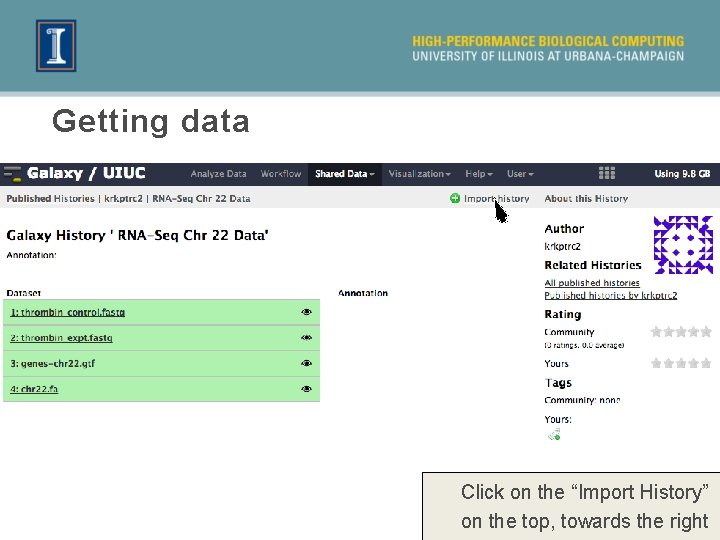
Getting data Click on the “Import History” on the top, towards the right

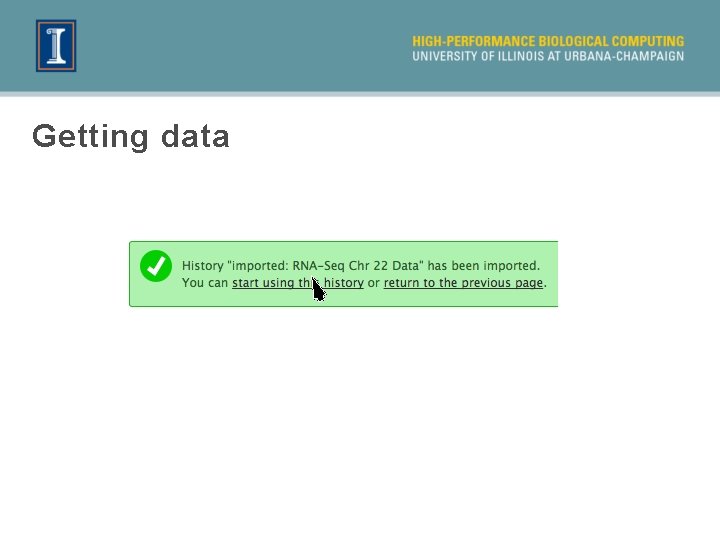
Getting data

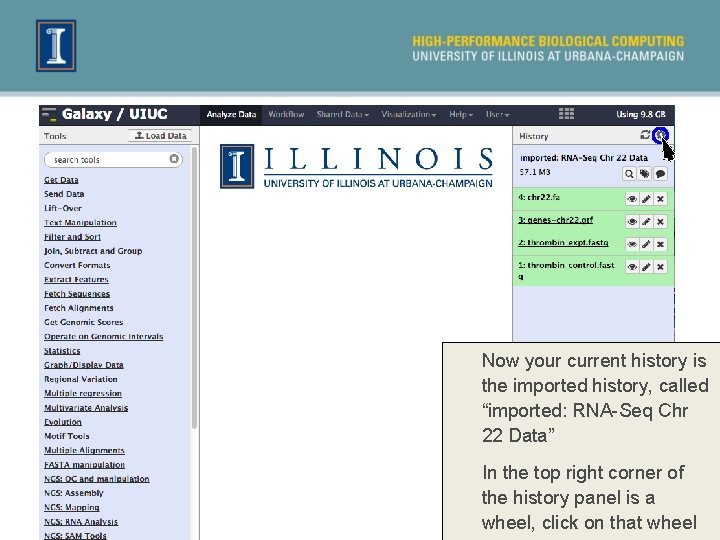
Now your current history is the imported history, called “imported: RNA-Seq Chr 22 Data” In the top right corner of the history panel is a wheel, click on that wheel

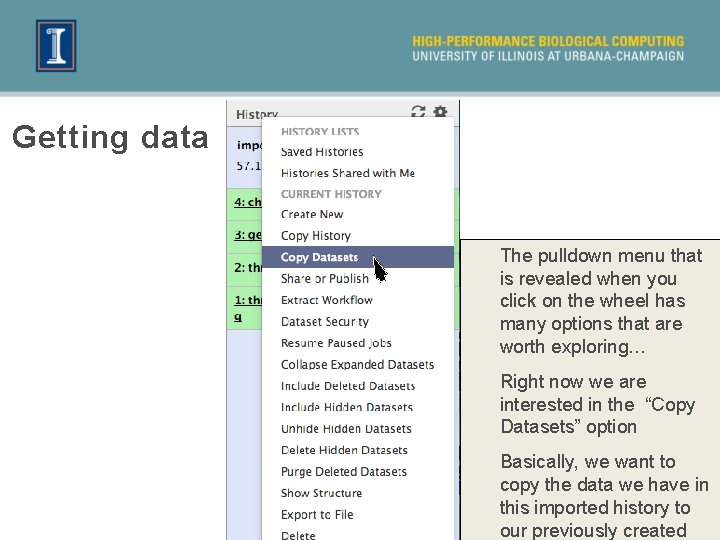
Getting data The pulldown menu that is revealed when you click on the wheel has many options that are worth exploring… Right now we are interested in the “Copy Datasets” option Basically, we want to copy the data we have in this imported history to our previously created

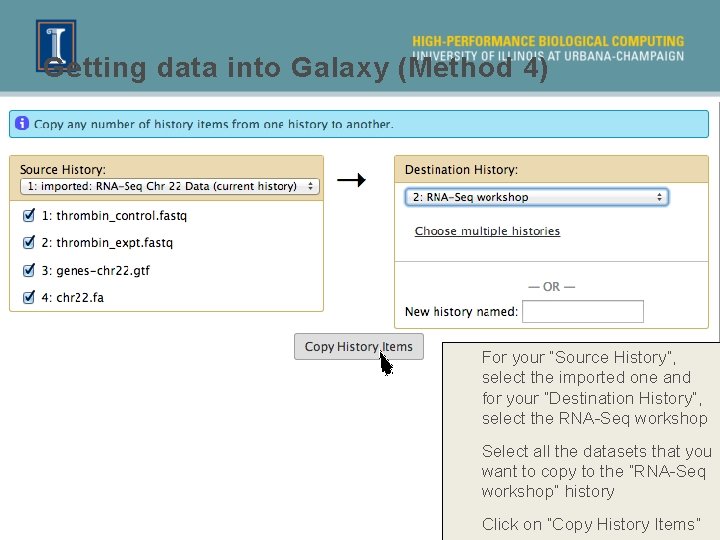
Getting data into Galaxy (Method 4) For your “Source History”, select the imported one and for your “Destination History”, select the RNA-Seq workshop Select all the datasets that you want to copy to the “RNA-Seq workshop” history Click on “Copy History Items”

Getting data

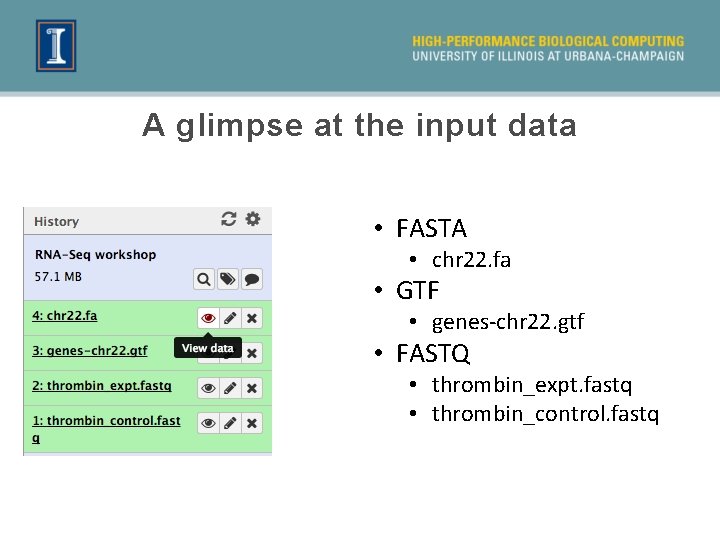
A glimpse at the input data • FASTA • chr 22. fa • GTF • genes-chr 22. gtf • FASTQ • thrombin_expt. fastq • thrombin_control. fastq

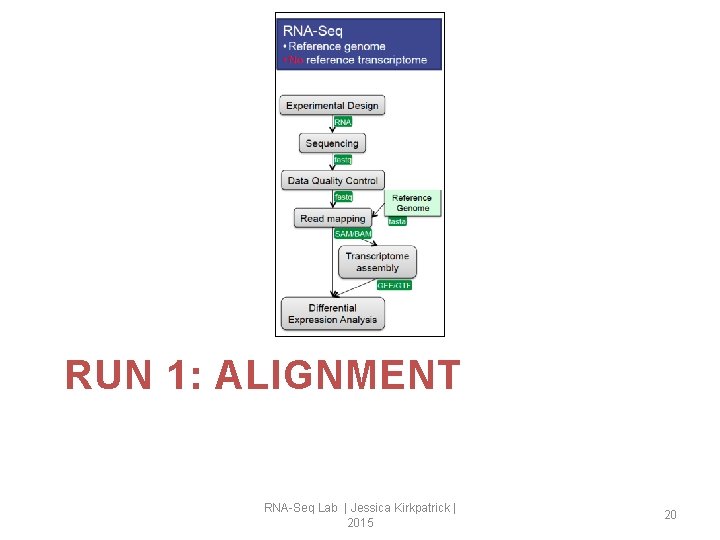
RUN 1: ALIGNMENT RNA-Seq Lab | Jessica Kirkpatrick | 2015 20

Aligning reads using Top. Hat We are not going to provide any genic structure information. Top. Hat will find splice junctions on its own. 21

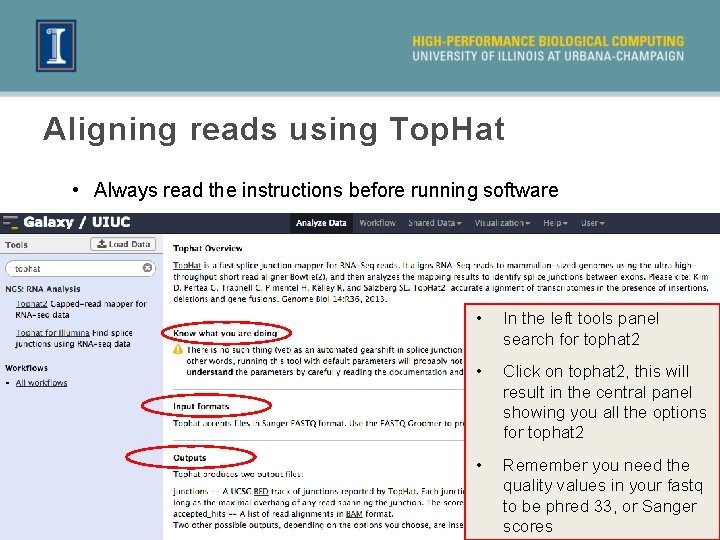
Aligning reads using Top. Hat • Always read the instructions before running software • In the left tools panel search for tophat 2 • Click on tophat 2, this will result in the central panel showing you all the options for tophat 2 • Remember you need the quality values in your fastq to be phred 33, or Sanger 22 scores

Aligning reads using Top. Hat 2 • Run 1: • No genic structure information (i. e. no GTF file) • Top. Hat 2 will find splice junctions on its own • Run this on experimental & control data. • Run 2: • Genic structure information will be used • Run this on experimental data. 23

Alignment with Tophat 2: Run 1 RNA-Seq Lab | Jessica Kirkpatrick | 2015 • In the left tools panel search for tophat 2 • Click on tophat 2, this will result in the central panel showing you all the options for tophat 2 • Remember you need the quality values in your fastq to be phred 33, or Sanger scores 24

Alignment with Tophat 2: Run 1 RNA-Seq Lab | Jessica Kirkpatrick | 2015 25

Alignment with Tophat 2: Run 1 RNA-Seq Lab | Jessica Kirkpatrick | 2015 26

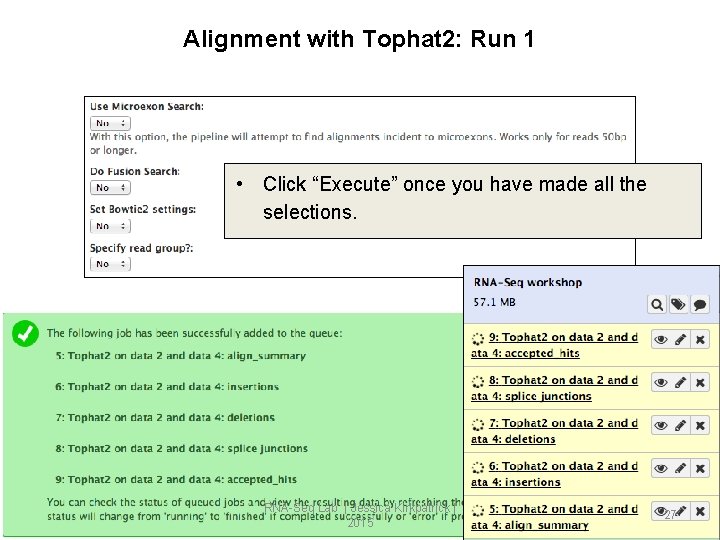
Alignment with Tophat 2: Run 1 • Click “Execute” once you have made all the selections. RNA-Seq Lab | Jessica Kirkpatrick | 2015 27

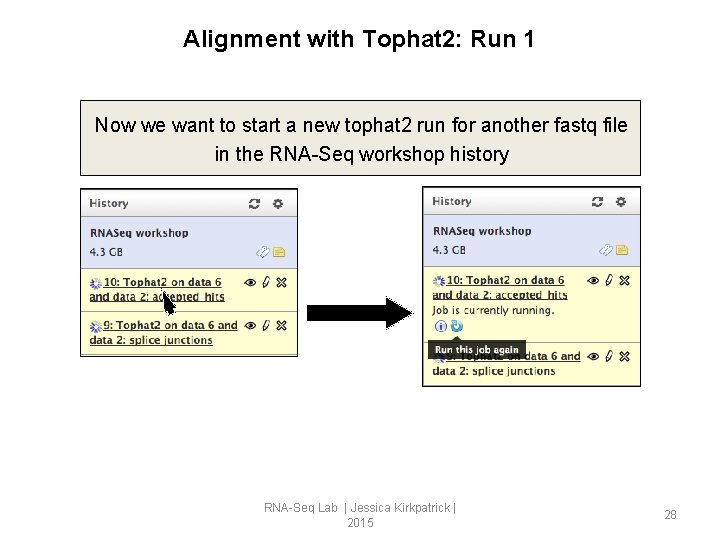
Alignment with Tophat 2: Run 1 Now we want to start a new tophat 2 run for another fastq file in the RNA-Seq workshop history RNA-Seq Lab | Jessica Kirkpatrick | 2015 28

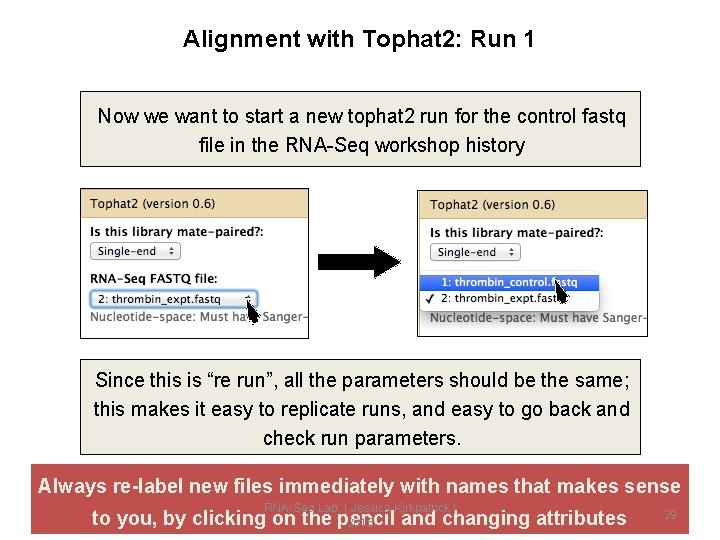
Alignment with Tophat 2: Run 1 Now we want to start a new tophat 2 run for the control fastq file in the RNA-Seq workshop history Since this is “re run”, all the parameters should be the same; this makes it easy to replicate runs, and easy to go back and check run parameters. Always re-label new files immediately with names that makes sense RNA-Seq Lab | Jessica Kirkpatrick | to you, by clicking on the pencil and changing attributes 2015 29

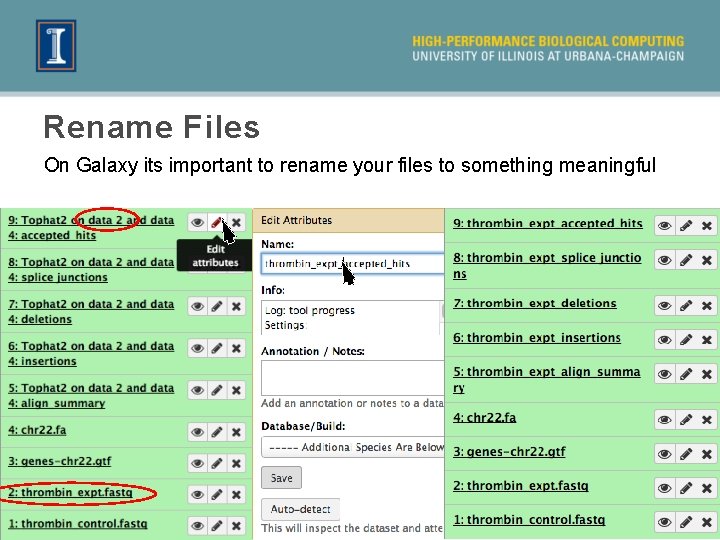
Rename Files On Galaxy its important to rename your files to something meaningful 30

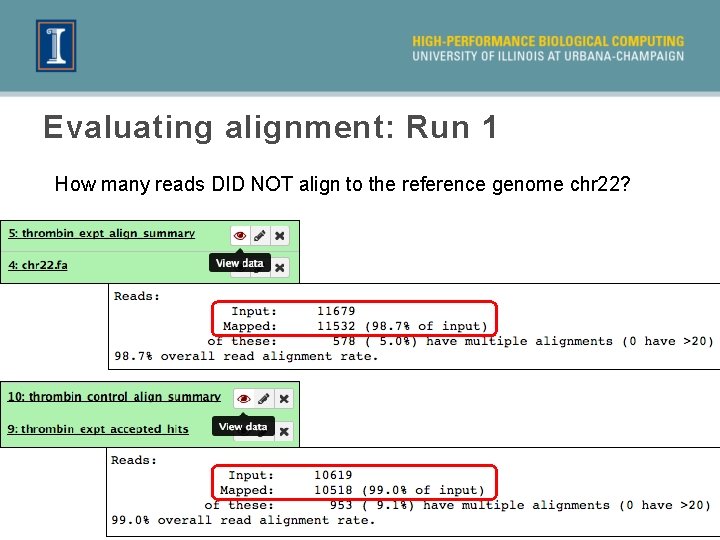
Evaluating alignment: Run 1 How many reads DID NOT align to the reference genome chr 22? 31

. RUN 2: INFORMED ALIGNMENT RNA-Seq Lab | Jessica Kirkpatrick | 2015 32

Aligning reads using Top. Hat 2 • Run 1: • No genic structure information (i. e. no GTF file) • Top. Hat 2 will find splice junctions on its own • Run this on experimental and control data • Run 2: • Genic structure information will be used • Run this on experimental data only 33

Alignment with Tophat 2: Run 2 Now we want to start a new informed tophat 2 run RNA-Seq Lab | Jessica Kirkpatrick | 2015 34

Aligning reads using gene information • Click “Execute” once you have changed the selections shown above. 35

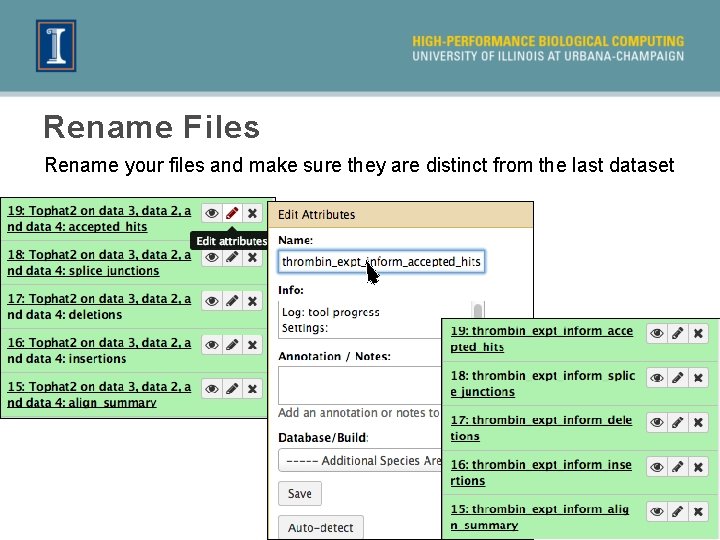
Rename Files Rename your files and make sure they are distinct from the last dataset 36

Evaluating alignment: Run 2 37

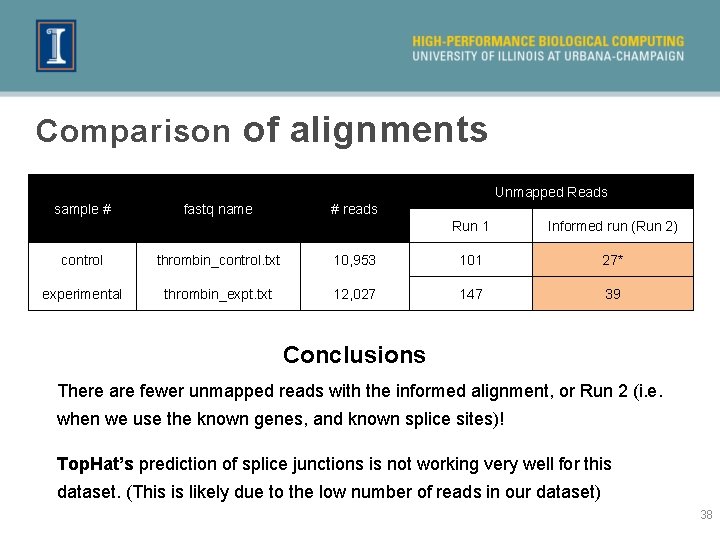
Comparison of alignments Unmapped Reads sample # fastq name # reads Run 1 Informed run (Run 2) control thrombin_control. txt 10, 953 101 27* experimental thrombin_expt. txt 12, 027 147 39 Conclusions There are fewer unmapped reads with the informed alignment, or Run 2 (i. e. when we use the known genes, and known splice sites)! Top. Hat’s prediction of splice junctions is not working very well for this dataset. (This is likely due to the low number of reads in our dataset) 38

. FINDING DIFFERENTIALLY EXPRESSED GENES RNA-Seq Lab | Jessica Kirkpatrick | 2015 39

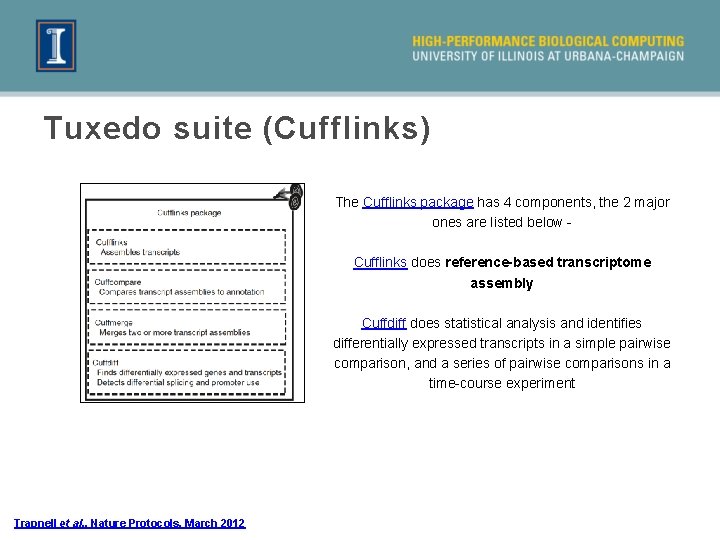
Tuxedo suite (Cufflinks) The Cufflinks package has 4 components, the 2 major ones are listed below Cufflinks does reference-based transcriptome assembly Cuffdiff does statistical analysis and identifies differentially expressed transcripts in a simple pairwise comparison, and a series of pairwise comparisons in a time-course experiment Trapnell et al. , Nature Protocols, March 2012

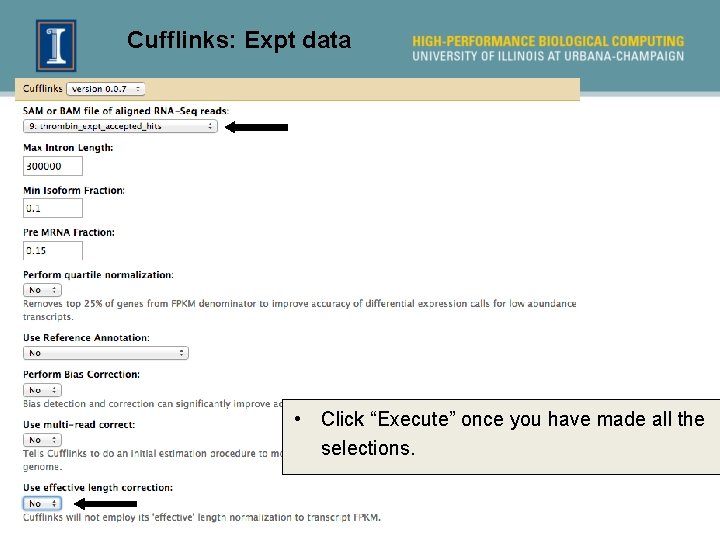
Assembling transcripts using Cufflinks • Run Cufflinks to obtain newly assembled gene transcripts from the aligned RNA-Seq reads. There is no need to conduct this step for the informed alignment (Run 2) because the locations of known genes are known already. 41

Cufflinks: Expt data • Click “Execute” once you have made all the selections.

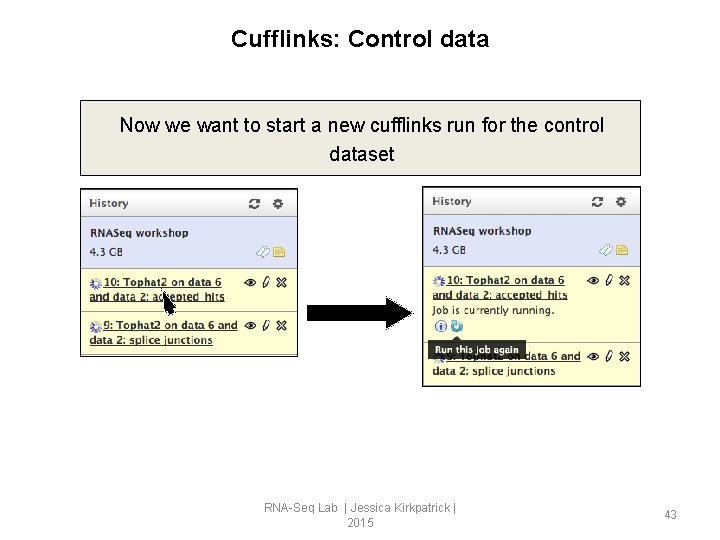
Cufflinks: Control data Now we want to start a new cufflinks run for the control dataset RNA-Seq Lab | Jessica Kirkpatrick | 2015 43

Cufflinks: Control data Now we want to start a new cufflinks run for the control dataset Since this is “re run”, all the parameters should be the same; this makes it easy to replicate runs, and easy to go back and check run parameters. RNA-Seq Lab | Jessica Kirkpatrick | 2015 44

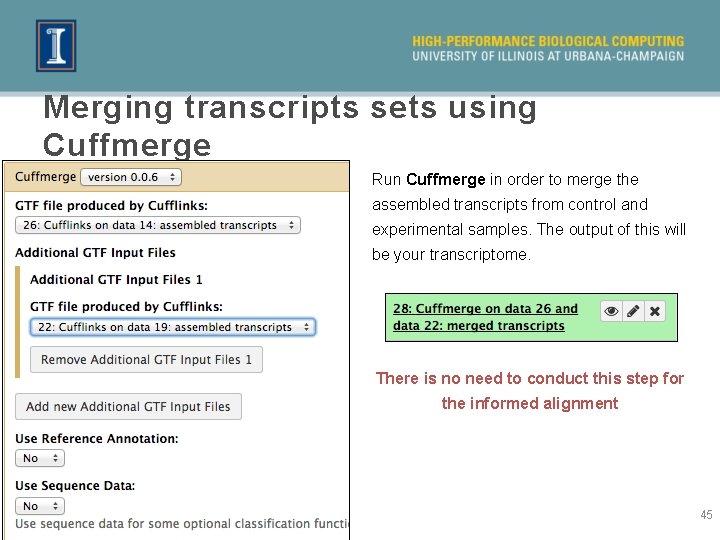
Merging transcripts sets using Cuffmerge Run Cuffmerge in order to merge the assembled transcripts from control and experimental samples. The output of this will be your transcriptome. There is no need to conduct this step for the informed alignment 45

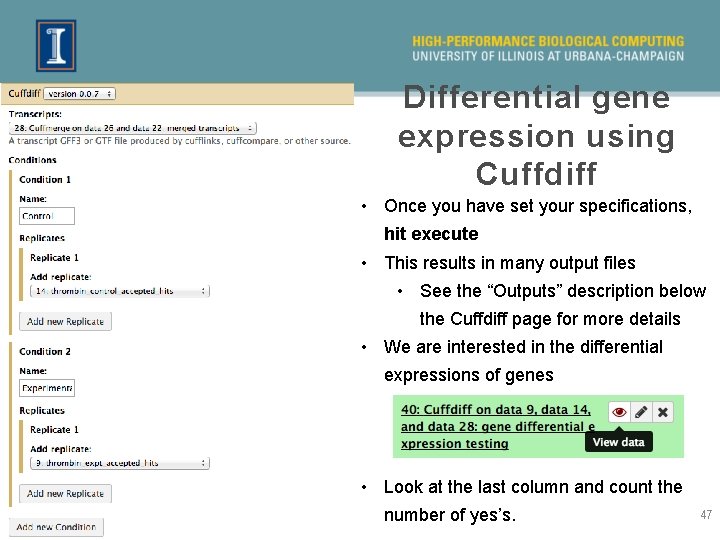
Differential gene expression using Cuffdiff • For Run 1 (uninformed) lets find out how many differentially expressed (DE) genes are present • We need a gene (. gtf) file and both the alignment (. bam) files (control and experimental) • We could use Cuffdiff on the informed alignments (run 2) as well, but we normally recommend using htseqcount and edge. R instead 46

Differential gene expression using Cuffdiff • Once you have set your specifications, hit execute • This results in many output files • See the “Outputs” description below the Cuffdiff page for more details • We are interested in the differential expressions of genes • Look at the last column and count the number of yes’s. 47

. VISUALIZATION USING IGV The Integrative Genomics Viewer (IGV) is a tool that supports the visualization of mapped reads to a reference genome, among other functionalities. RNA-Seq Lab | Jessica Kirkpatrick | 2015 48

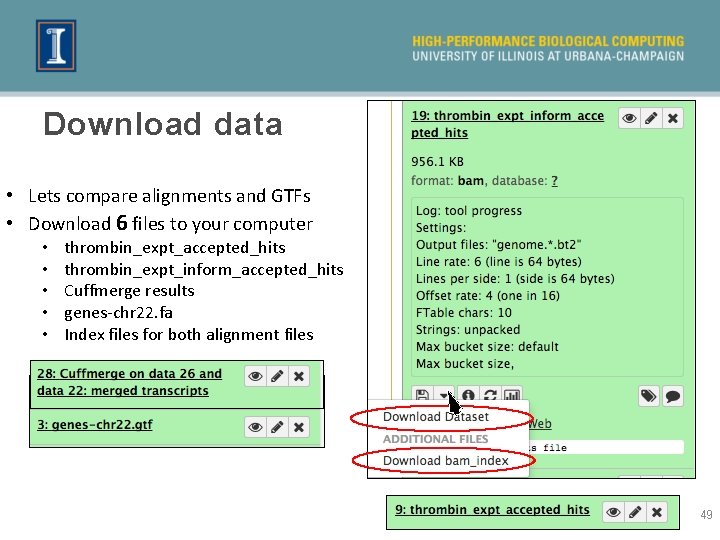
Download data • Lets compare alignments and GTFs • Download 6 files to your computer • • • thrombin_expt_accepted_hits thrombin_expt_inform_accepted_hits Cuffmerge results genes-chr 22. fa Index files for both alignment files 49

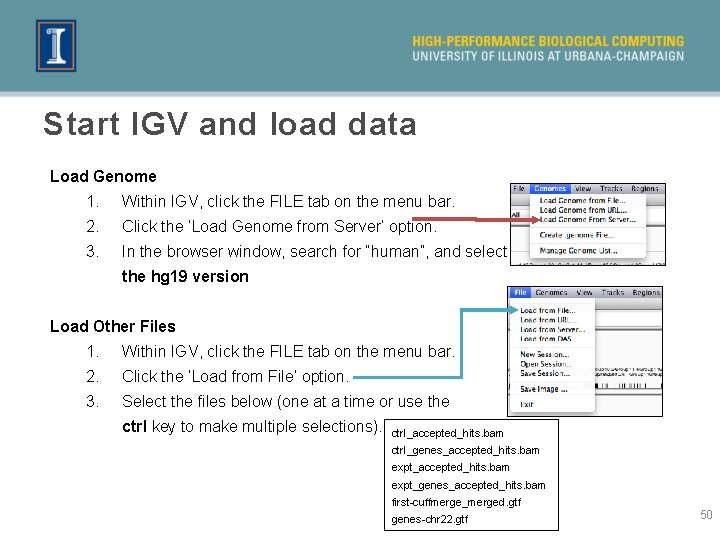
Start IGV and load data Load Genome 1. Within IGV, click the FILE tab on the menu bar. 2. Click the ‘Load Genome from Server’ option. 3. In the browser window, search for “human”, and select the hg 19 version Load Other Files 1. Within IGV, click the FILE tab on the menu bar. 2. Click the ‘Load from File’ option. 3. Select the files below (one at a time or use the ctrl key to make multiple selections). ctrl_accepted_hits. bam ctrl_genes_accepted_hits. bam expt_genes_accepted_hits. bam first-cuffmerge_merged. gtf genes-chr 22. gtf 50

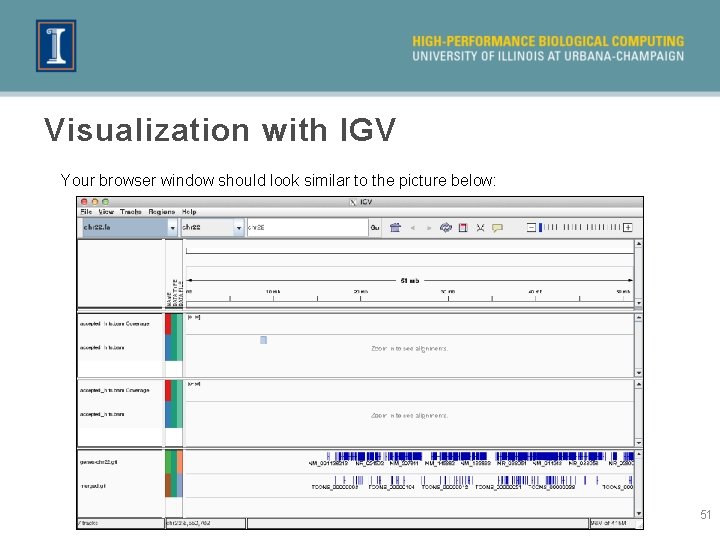
Visualization with IGV Your browser window should look similar to the picture below: 51

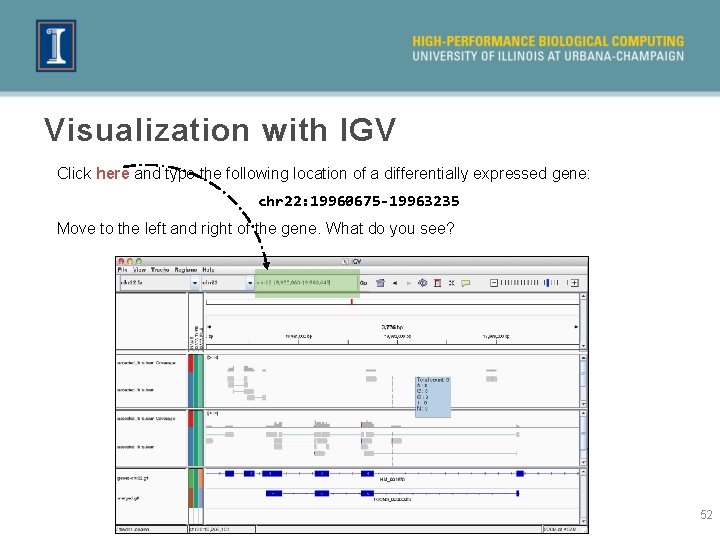
Visualization with IGV Click here and type the following location of a differentially expressed gene: chr 22: 19960675 -19963235 Move to the left and right of the gene. What do you see? 52

Visualization with IGV » Looks like the new transcriptome (first-cuffmerge_merged. gtf) compares poorly to the known gene models. This is very likely due to the very low number of reads in our dataset. » We can see that there are many more reads for one dataset compared to the other. Hence, it makes sense that the gene was called as being differentially expressed. » Note the intron spanning reads. 53

Conclusion Today we did the following: 1. Used the Tuxedo Suite to: a. Aligned RNA-Seq reads using Top. Hat(splice-aware aligner). b. Performed reference-based transcriptome assembly with Cufflinks. c. Obtained a new transcriptome using Cufflinks & Cuffmerge. d. Used Cuffdiff to obtain a list of differentially expressed genes. e. Reported a list of significantly expressed genes. 2. Used a genome browser and visualization tool to observe the aligned data and the new transcriptome. 54

Useful links Online resources for RNA-Seq analysis questions – ² http: //www. biostars. org/ - Biostar (Bioinformatics explained) ² http: //seqanswers. com/ - SEQanswers (the next generation sequencing community) ² Most tools have a dedicated lists Information about the various parts of the Tuxedo suite is available here http: //ccb. jhu. edu/software. shtml Genome Browsers tutorials – ² http: //www. broadinstitute. org/igv/Quick. Start/ - IGV tutorials ² http: //www. openhelix. com/ucsc/ - UCSC browser tutorials (openhelix is a great place for tutorials, UIUC has a campus-wide subscription) Contact us at: hpcbiohelp@illinois. edu hpcbiotraining@illinois. edu 55