Identification and Structure Determination of Higher Order Glycosphingolipids








- Slides: 33

Identification and Structure Determination of Higher Order Glycosphingolipids via LC-MS/MS M. Cameron Sullards, Ph. D. Georgia Institute of Technology: School of Chemistry and Biochemistry and School of Biology, Atlanta, GA 30332 -0363

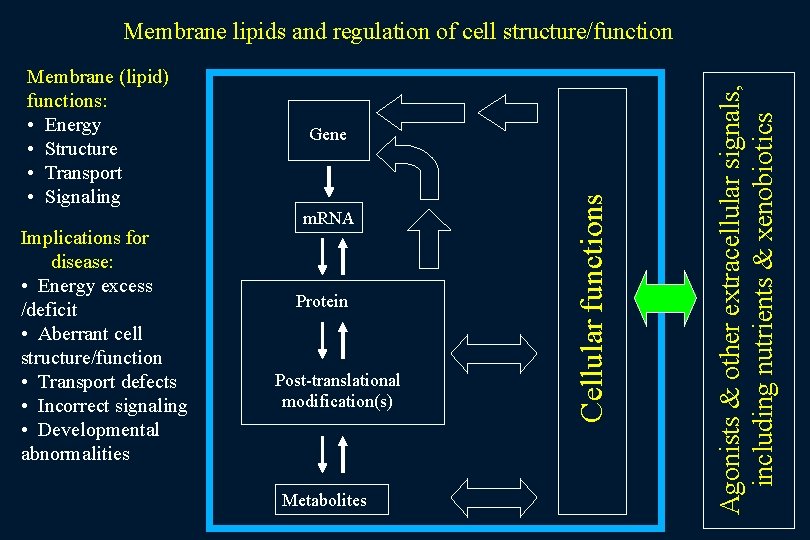
Implications for disease: • Energy excess /deficit • Aberrant cell structure/function • Transport defects • Incorrect signaling • Developmental abnormalities Gene m. RNA Protein Post-translational modification(s) Metabolites Cellular functions Membrane (lipid) functions: • Energy • Structure • Transport • Signaling Agonists & other extracellular signals, including nutrients & xenobiotics Membrane lipids and regulation of cell structure/function

LIPID MAPS

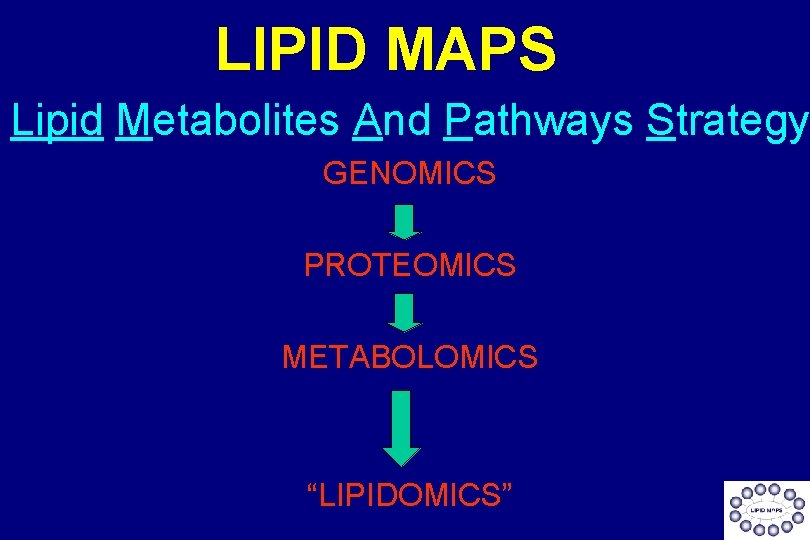
LIPID MAPS Lipid Metabolites And Pathways Strategy GENOMICS PROTEOMICS METABOLOMICS “LIPIDOMICS”

Lipid Metabolites and Pathways Strategy LIPID MAPS GOALS (1) To separate and detect all of the lipids in a specific cell and to discover and characterize any novel lipids that may be present. (2) To quantitate each of the lipid metabolites present and to quantitate the changes in their levels and location during cellular function. (3) To define the biochemical pathways for each lipid and develop lipid maps which define the interaction networks.

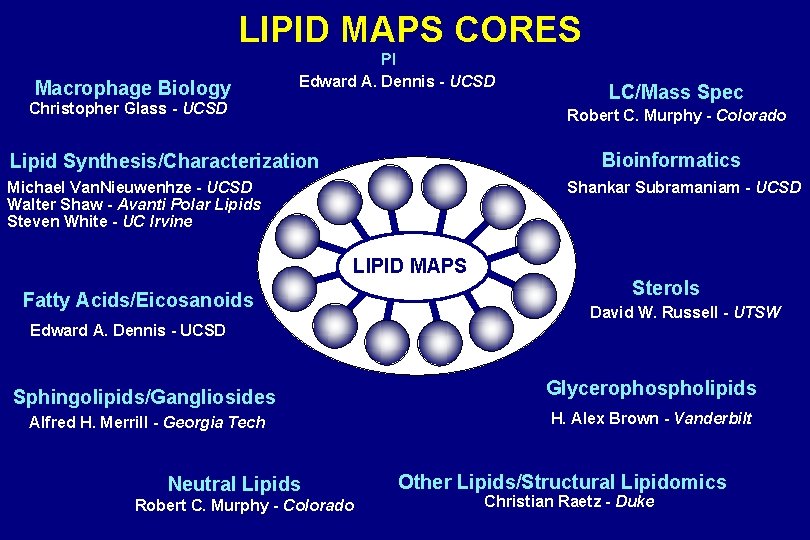
LIPID MAPS CORES Macrophage Biology PI Edward A. Dennis - UCSD Christopher Glass - UCSD LC/Mass Spec Robert C. Murphy - Colorado Bioinformatics Lipid Synthesis/Characterization Michael Van. Nieuwenhze - UCSD Walter Shaw - Avanti Polar Lipids Steven White - UC Irvine Shankar Subramaniam - UCSD LIPID MAPS Fatty Acids/Eicosanoids Edward A. Dennis - UCSD Sphingolipids/Gangliosides Alfred H. Merrill - Georgia Tech Neutral Lipids Robert C. Murphy - Colorado Sterols David W. Russell - UTSW Glycerophospholipids H. Alex Brown - Vanderbilt Other Lipids/Structural Lipidomics Christian Raetz - Duke

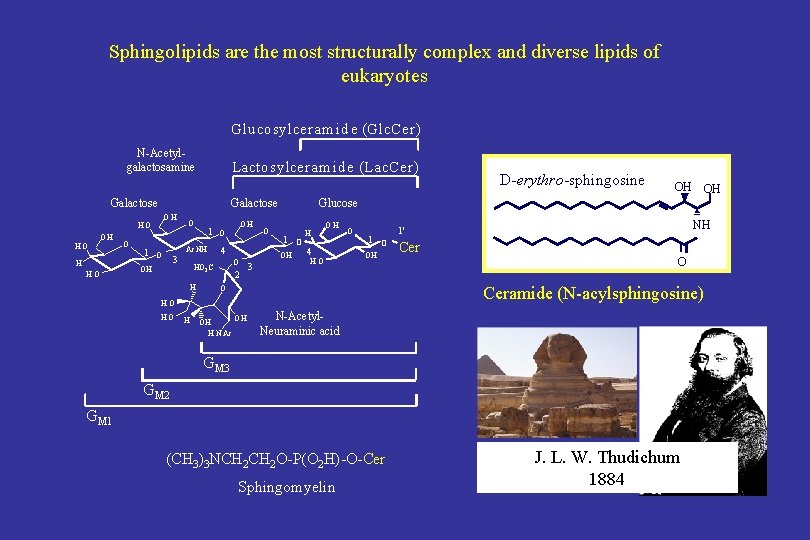
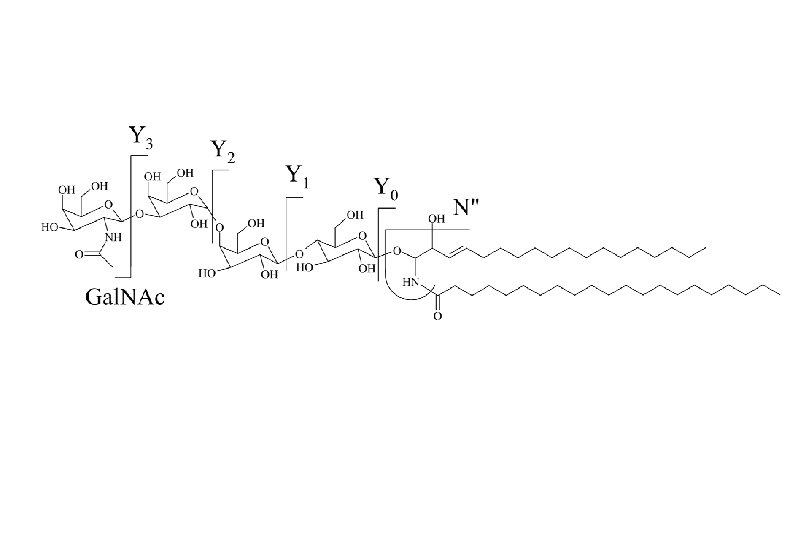
Sphingolipids are the most structurally complex and diverse lipids of eukaryotes Glu cosylceram id e (Glc. Cer) N-Acetylgalactosamine Lactosylcera m id e (La c. Cer) Galactose OH HO O 1 Ac. NH O 3 OH O 4 O HO 2 C OH H 2 O 1 O H OH 4 HO O 1 O OH H NH 1' Cer O Ceramide (N-acylsphingosine) O HO HO OH OH Glucose OH 3 D-erythro-sphingosine OH OH H N Ac N-Acetyl. Neuraminic acid GM 3 GM 2 GM 1 (CH 3)3 NCH 2 O-P(O 2 H)-O-Cer Sphingomyelin J. L. W. Thudichum 1884

Sphin. GOMAP© (Download available at www. sphingomap. org)

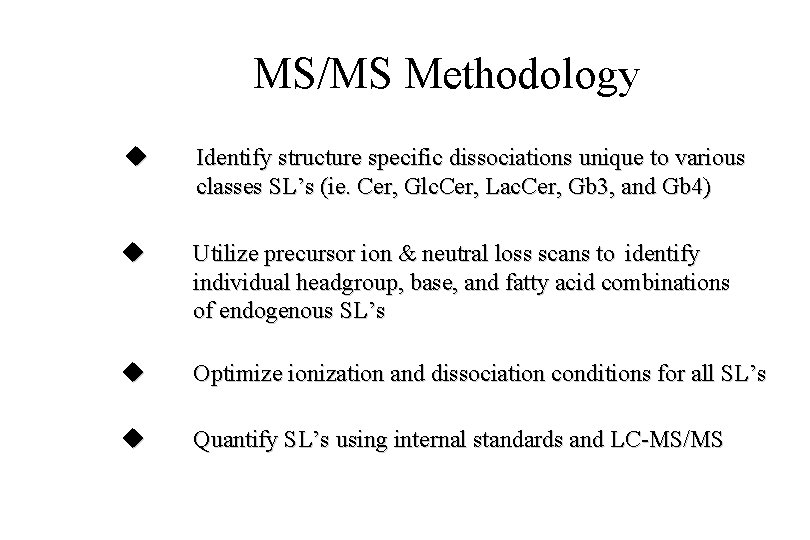
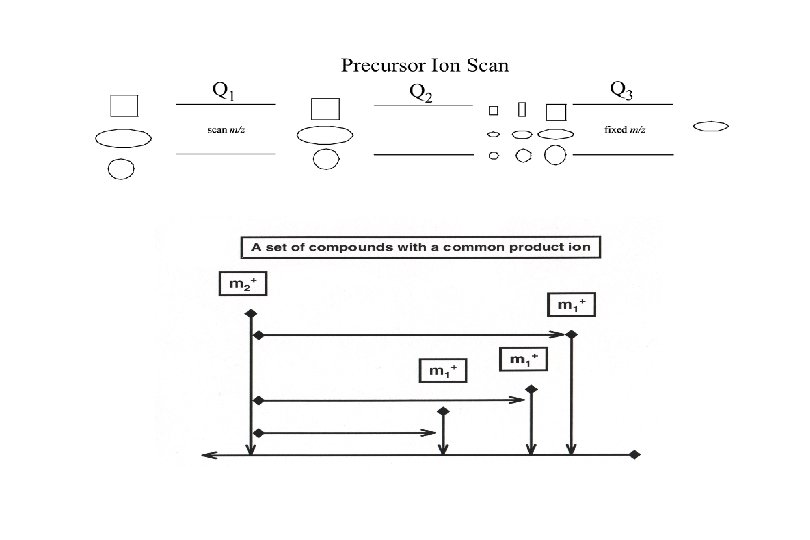
MS/MS Methodology Identify structure specific dissociations unique to various classes SL’s (ie. Cer, Glc. Cer, Lac. Cer, Gb 3, and Gb 4) Utilize precursor ion & neutral loss scans to identify individual headgroup, base, and fatty acid combinations of endogenous SL’s Optimize ionization and dissociation conditions for all SL’s Quantify SL’s using internal standards and LC-MS/MS

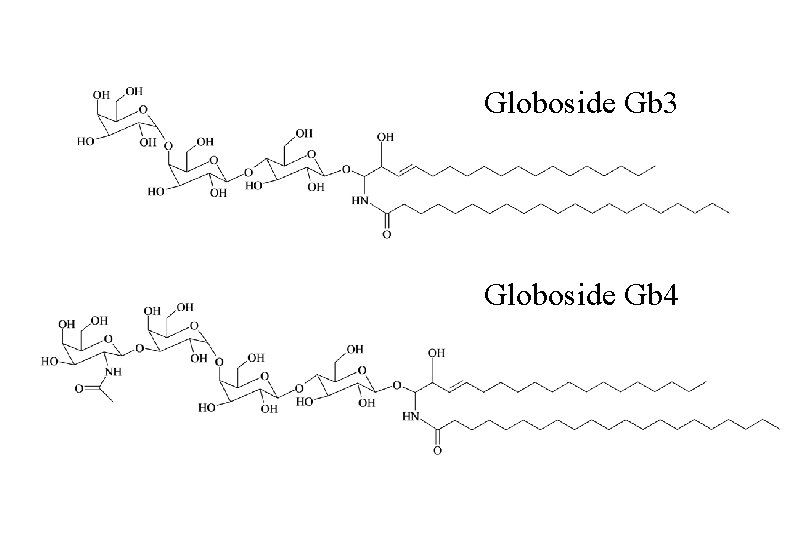
Globoside Gb 3 Globoside Gb 4


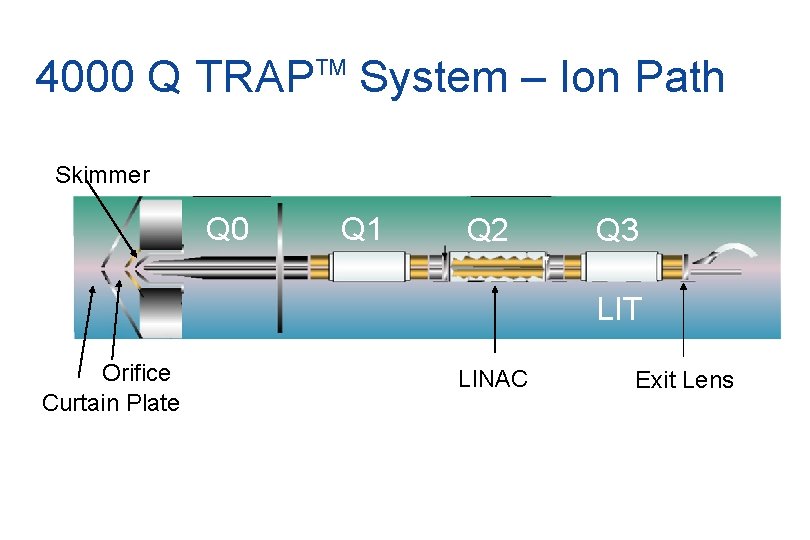
4000 Q TRAP System – Ion Path TM Skimmer Q 0 Q 1 Q 2 Q 3 LIT Orifice Curtain Plate LINAC Exit Lens

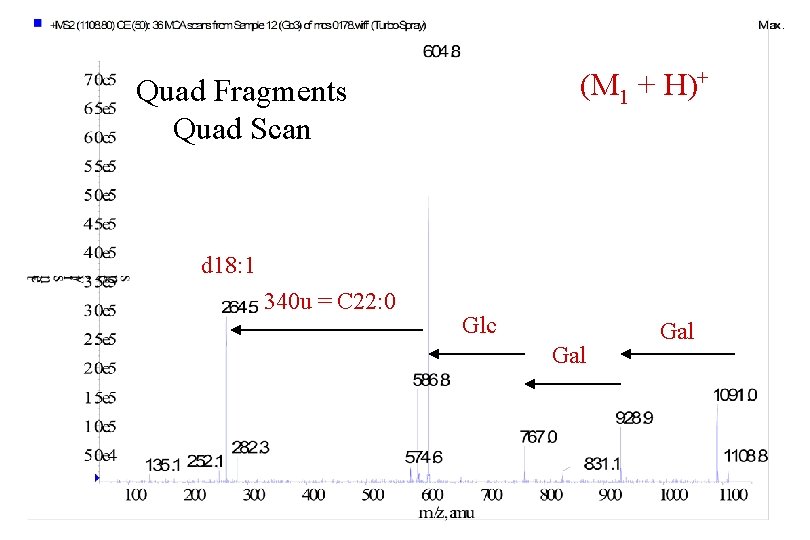
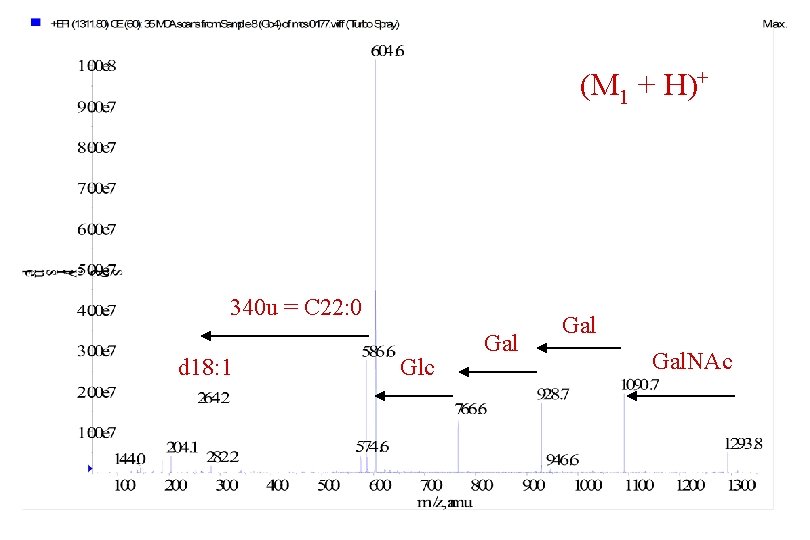
(M 1 + H)+ Quad Fragments Quad Scan d 18: 1 340 u = C 22: 0 Glc Gal


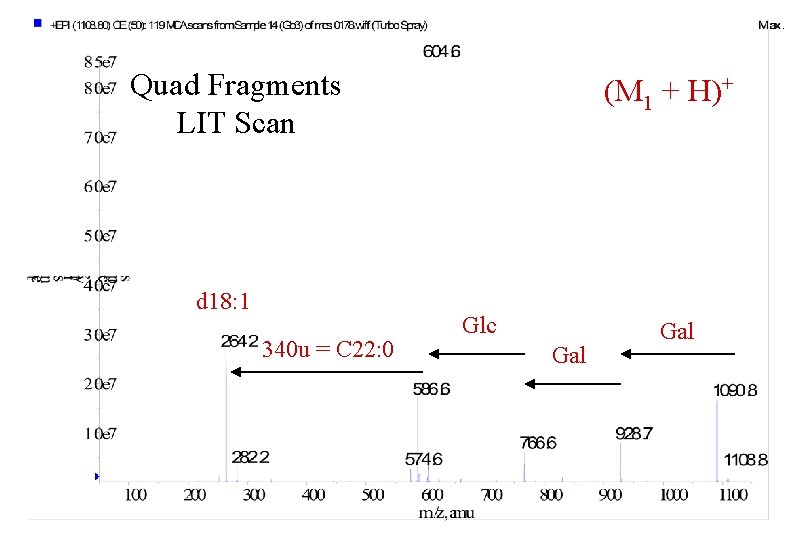
Quad Fragments LIT Scan d 18: 1 340 u = C 22: 0 (M 1 + H)+ Glc Gal

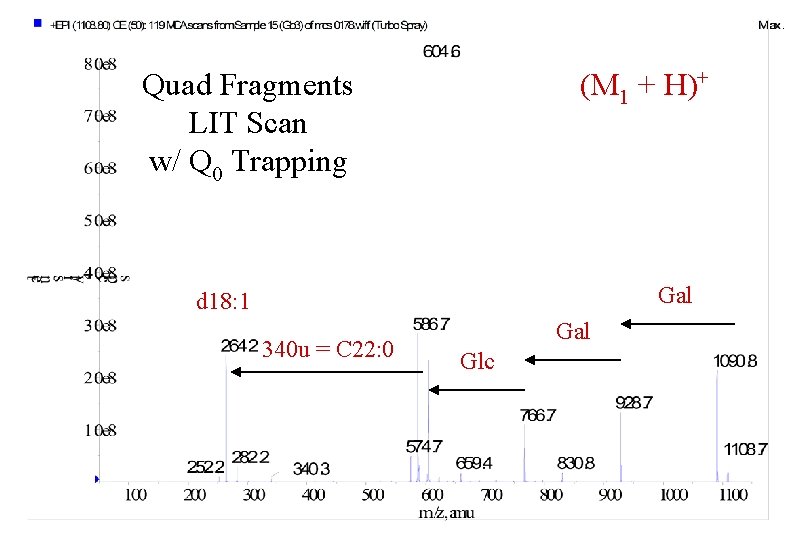
Quad Fragments LIT Scan w/ Q 0 Trapping (M 1 + H)+ Gal d 18: 1 340 u = C 22: 0 Gal Glc

4000 Q TRAP System – Ion Path TM Skimmer Q 0 Q 1 Q 2 Q 3 LIT Orifice Curtain Plate LINAC Exit Lens

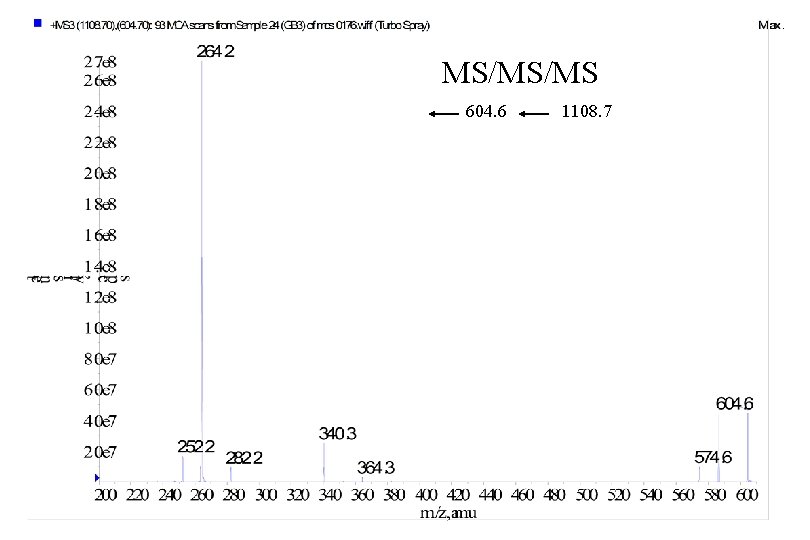
MS/MS/MS 604. 6 1108. 7

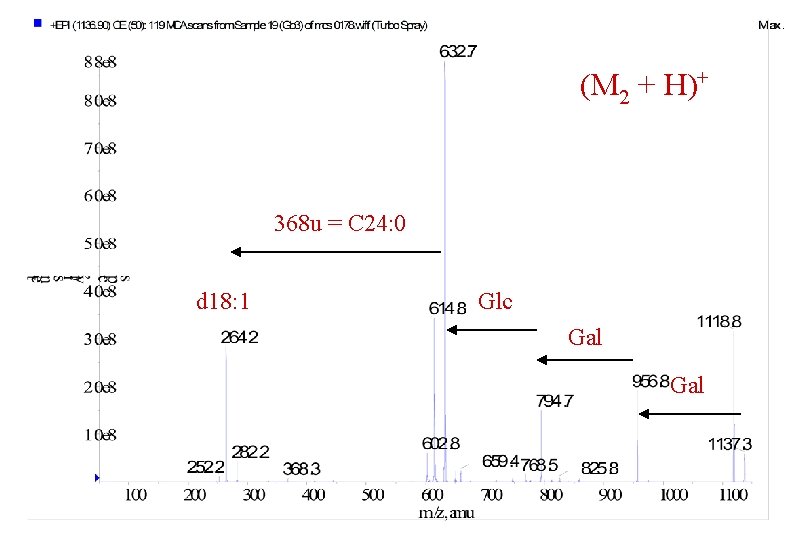
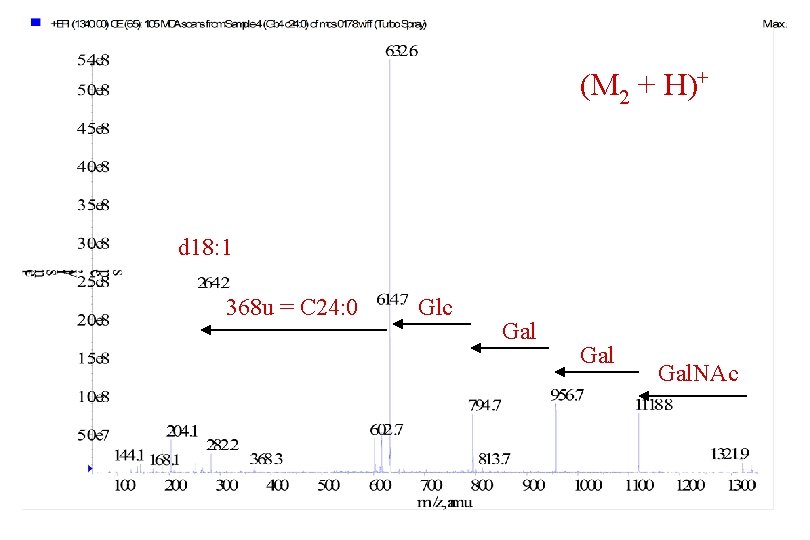
(M 2 + H)+ 368 u = C 24: 0 d 18: 1 Glc Gal

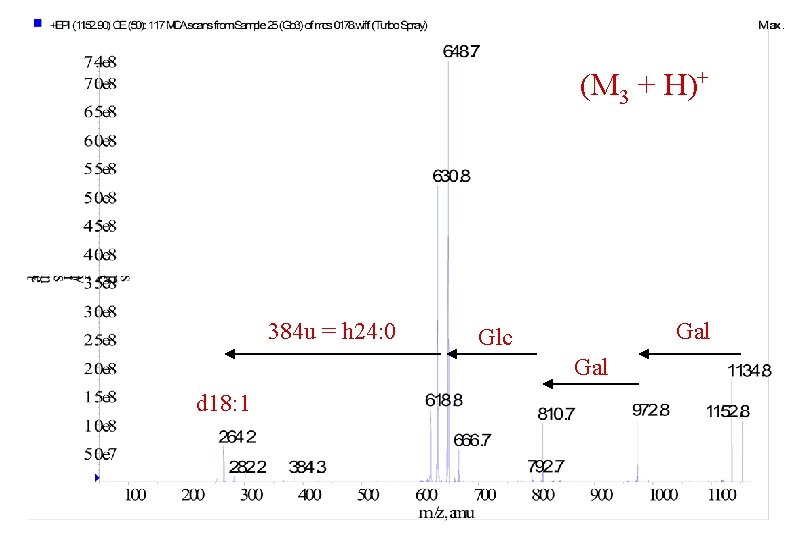
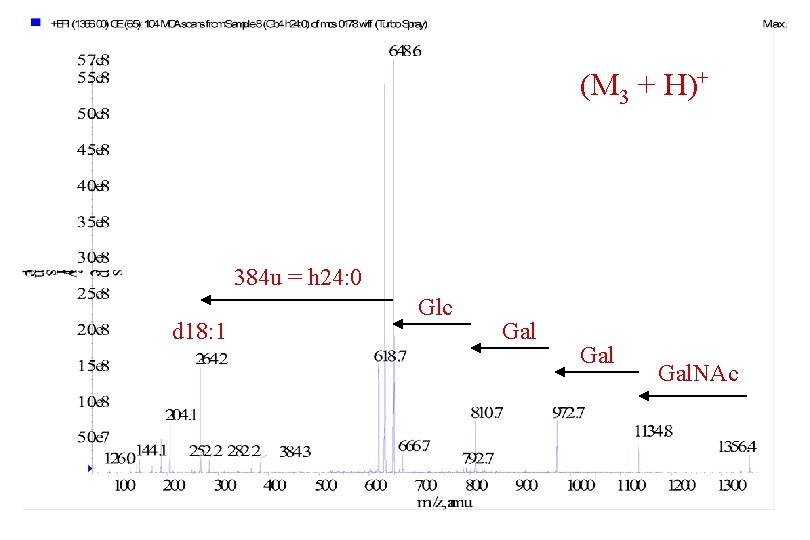
(M 3 + H)+ 384 u = h 24: 0 Gal Glc Gal d 18: 1


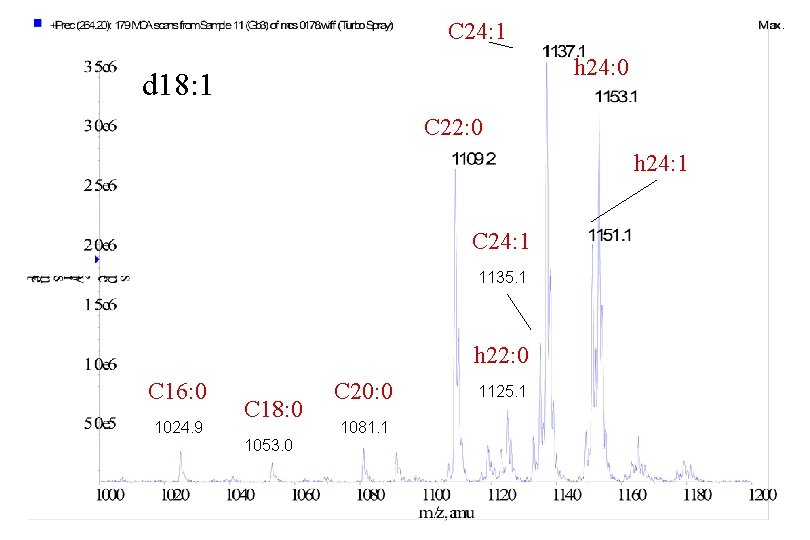
C 24: 1 h 24: 0 d 18: 1 C 22: 0 h 24: 1 C 24: 1 1135. 1 h 22: 0 C 16: 0 1024. 9 C 18: 0 1053. 0 C 20: 0 1081. 1 1125. 1



(M 1 + H)+ 340 u = C 22: 0 d 18: 1 Glc Gal Gal. NAc

(M 2 + H)+ d 18: 1 368 u = C 24: 0 Glc Gal Gal. NAc

(M 3 + H)+ 384 u = h 24: 0 d 18: 1 Glc Gal Gal. NAc

C 24: 0 d 18: 1 h 24: 0 C 22: 0 h 24: 1 C 16: 0 1228. 0 C 18: 0 C 20: 0 1256. 1 1284. 1

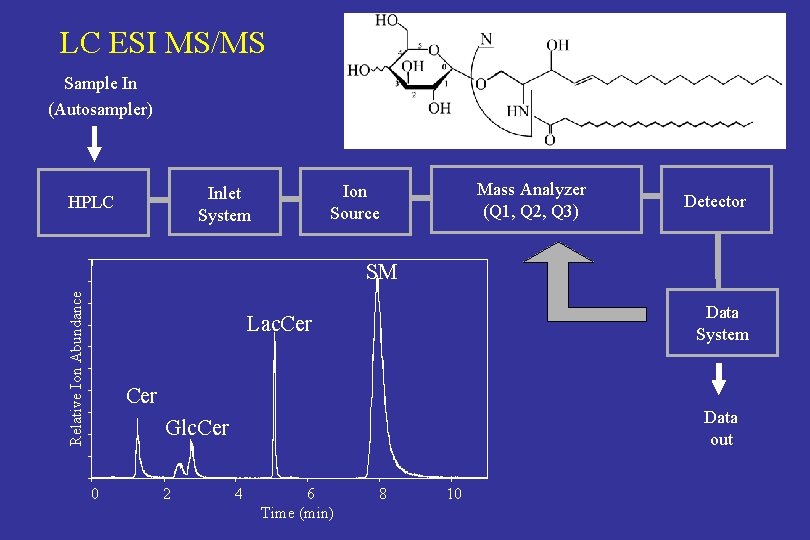
LC ESI MS/MS Sample In (Autosampler) Mass Analyzer (Q 1, Q 2, Q 3) Ion Source Inlet System HPLC Detector Relative Ion Abundance SM Data System Lac. Cer Data out Glc. Cer 0 2 4 6 Time (min) 8 10

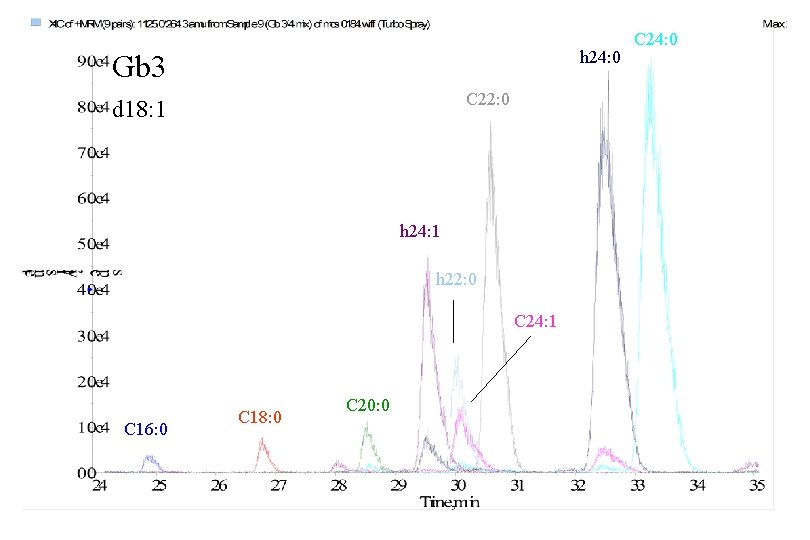
h 24: 0 Gb 3 C 22: 0 d 18: 1 h 24: 1 h 22: 0 C 24: 1 C 16: 0 C 18: 0 C 20: 0 C 24: 0

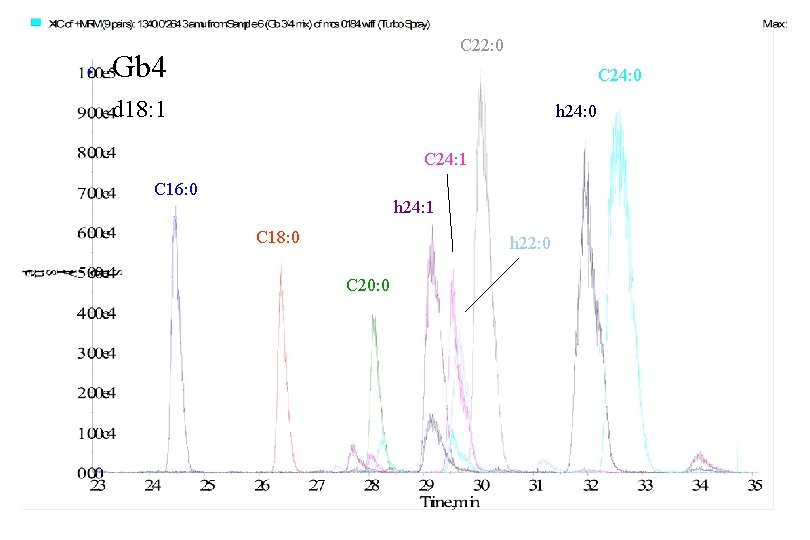
C 22: 0 Gb 4 C 24: 0 d 18: 1 h 24: 0 C 24: 1 C 16: 0 h 24: 1 C 18: 0 h 22: 0 C 20: 0

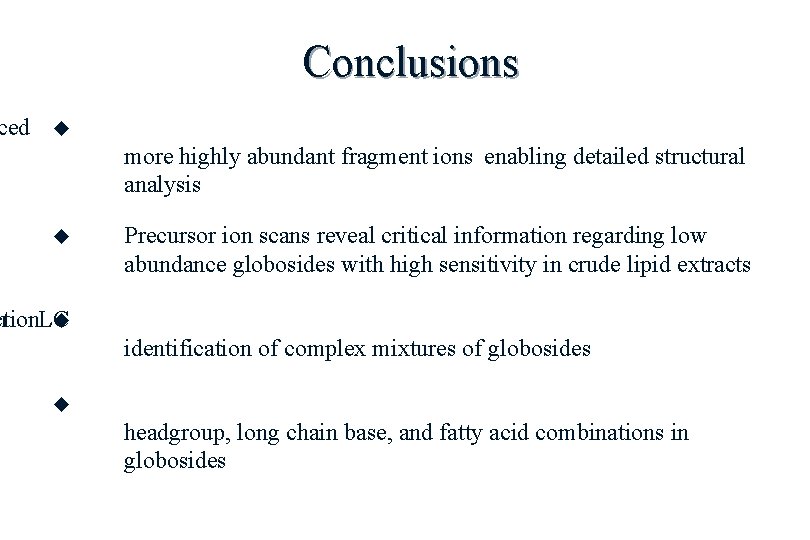
ced Conclusions more highly abundant fragment ions enabling detailed structural analysis Precursor ion scans reveal critical information regarding low abundance globosides with high sensitivity in crude lipid extracts ction n LC identification of complex mixtures of globosides headgroup, long chain base, and fatty acid combinations in globosides

Acknowledgements Prof. Alfred H. Merrill, Jr. Meeyoung Park Anu Koppikar http: //www. sphingomap. org Matreya NIH/NIGMS/Lipid MAPS http: //www. lipidmaps. org
 What is the function of glycolipids in the plasma membrane
What is the function of glycolipids in the plasma membrane Higher order structure of proteins
Higher order structure of proteins Central pocket whorl vs plain whorl
Central pocket whorl vs plain whorl Via optica
Via optica Product and quotient rules and higher order derivatives
Product and quotient rules and higher order derivatives X-ray structure determination
X-ray structure determination Anomalous
Anomalous Higher order function haskell
Higher order function haskell Higher order thinking skills
Higher order thinking skills Psychomotor objectives verbs
Psychomotor objectives verbs Higher order thinking iteach flashcards
Higher order thinking iteach flashcards Std bind
Std bind Higher order derivatives meaning
Higher order derivatives meaning Higher-order systems
Higher-order systems Hots questions examples
Hots questions examples Factoring higher degree polynomials
Factoring higher degree polynomials Contoh turunan parsial pertama
Contoh turunan parsial pertama Higher order functions haskell
Higher order functions haskell Extinction ap psychology
Extinction ap psychology Higher order thinking questions for vocabulary
Higher order thinking questions for vocabulary Bloom's taxonomy metacognition
Bloom's taxonomy metacognition Higher order linear differential equations
Higher order linear differential equations Higher
Higher Higher order functions haskell
Higher order functions haskell Haskell higher order functions
Haskell higher order functions Higher order derivatives
Higher order derivatives Definition of higher order conditioning
Definition of higher order conditioning Maniac magee higher order thinking questions
Maniac magee higher order thinking questions Learned helplessness ap psychology definition
Learned helplessness ap psychology definition Y chromosome traits
Y chromosome traits Sex linkage
Sex linkage Quality control standards
Quality control standards Price and output determination under oligopoly
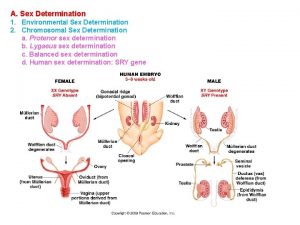
Price and output determination under oligopoly Sex determination and sex linkage
Sex determination and sex linkage