Genetics tools Next Generation Sequencing Illumina Hi Seq









- Slides: 19

Genetics tools – Next Generation Sequencing Illumina Hi. Seq Pac. Bio Nanopore

Considerations • Sequencing libraries • Amplification • Sequence by synthesis • Short reads • Data analysis • Sequence assembly Vocabulary • Adapter • Bar code • Bridge amplification • Cluster • Flow cell • Lanes • Multiplexing • Paired end reads • Reversible terminator Illumina Hi. Seq – short read example

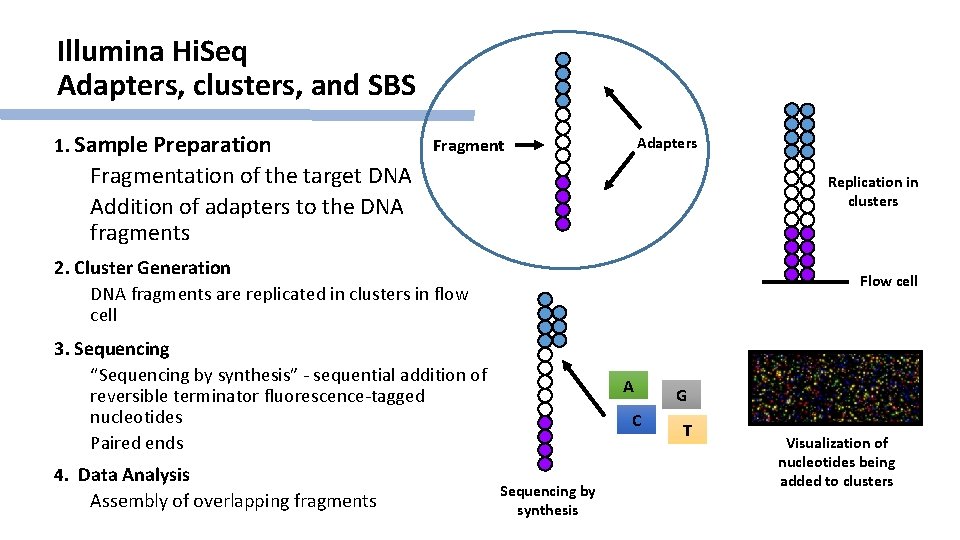
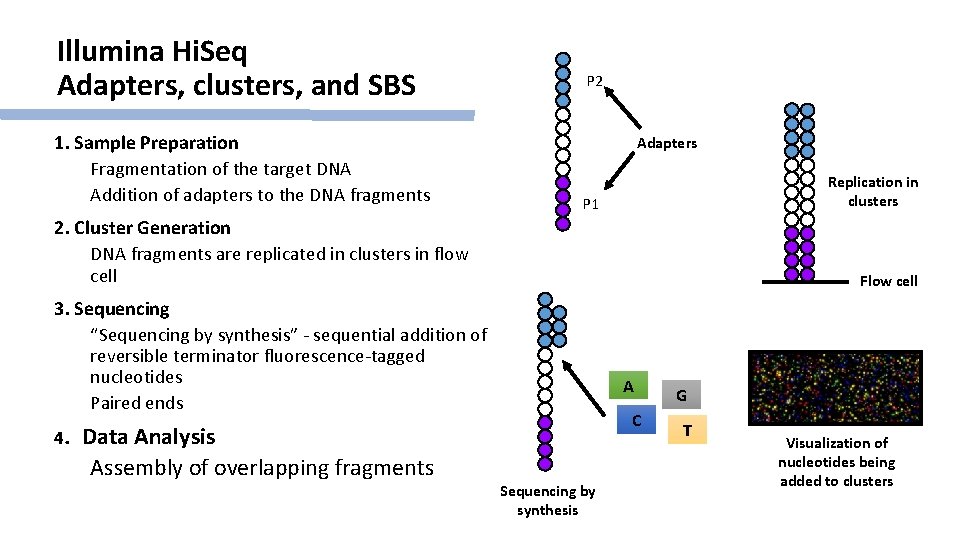
Illumina Hi. Seq Adapters, clusters, and SBS 1. Sample Preparation Adapters Fragmentation of the target DNA Addition of adapters to the DNA fragments Replication in clusters 2. Cluster Generation DNA fragments are replicated in clusters in flow cell Flow cell 3. Sequencing “Sequencing by synthesis” - sequential addition of reversible terminator fluorescence-tagged nucleotides Paired ends 4. Data Analysis Assembly of overlapping fragments A C Sequencing by synthesis G T Visualization of nucleotides being added to clusters

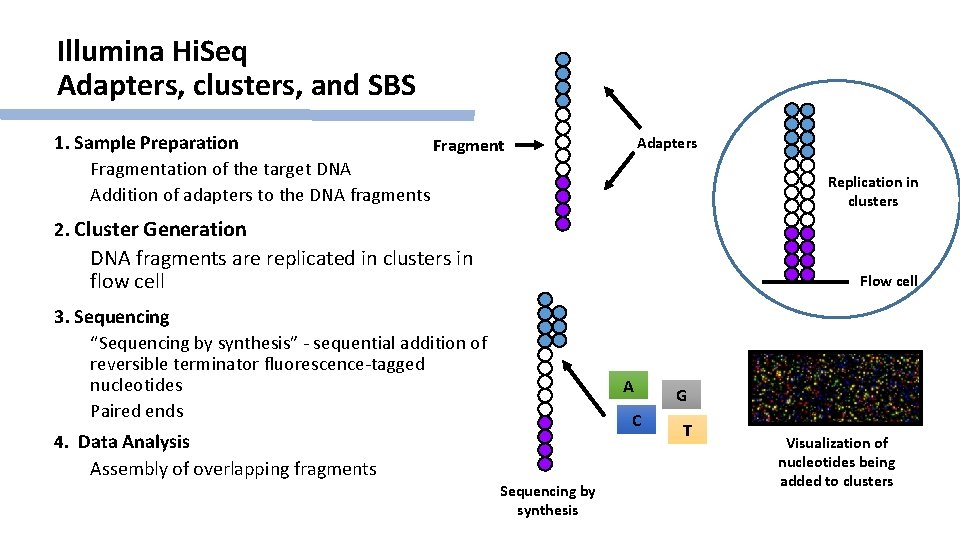
Illumina Hi. Seq Adapters, clusters, and SBS 1. Sample Preparation Fragmentation of the target DNA Addition of adapters to the DNA fragments Adapters Replication in clusters 2. Cluster Generation DNA fragments are replicated in clusters in flow cell Flow cell 3. Sequencing “Sequencing by synthesis” - sequential addition of reversible terminator fluorescence-tagged nucleotides Paired ends A C 4. Data Analysis Assembly of overlapping fragments Sequencing by synthesis G T Visualization of nucleotides being added to clusters

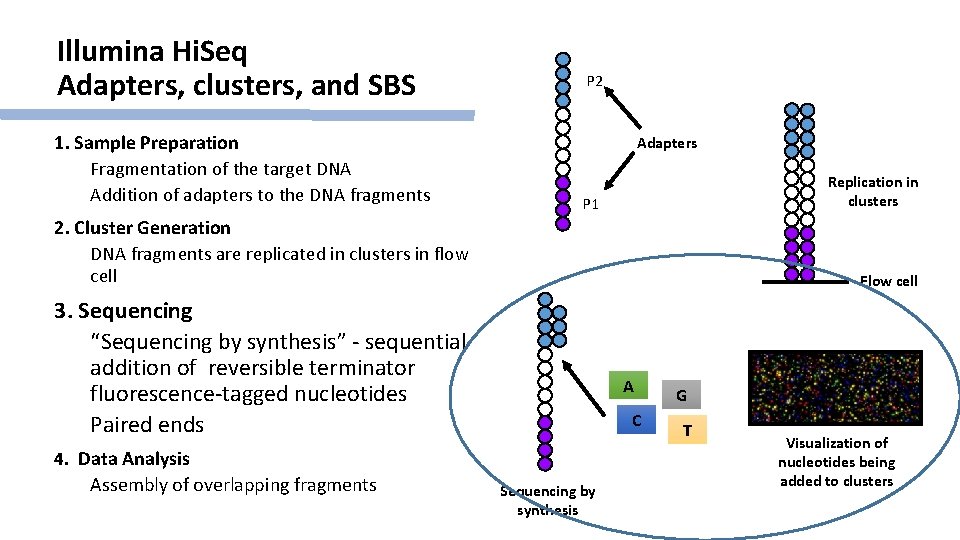
Illumina Hi. Seq Adapters, clusters, and SBS 1. Sample Preparation Fragmentation of the target DNA Addition of adapters to the DNA fragments P 2 Adapters Replication in clusters P 1 2. Cluster Generation DNA fragments are replicated in clusters in flow cell Flow cell 3. Sequencing “Sequencing by synthesis” - sequential addition of reversible terminator fluorescence-tagged nucleotides Paired ends 4. Data Analysis Assembly of overlapping fragments A C Sequencing by synthesis G T Visualization of nucleotides being added to clusters

Illumina Hi. Seq Adapters, clusters, and SBS 1. Sample Preparation Fragmentation of the target DNA Addition of adapters to the DNA fragments P 2 Adapters Replication in clusters P 1 2. Cluster Generation DNA fragments are replicated in clusters in flow cell Flow cell 3. Sequencing “Sequencing by synthesis” - sequential addition of reversible terminator fluorescence-tagged nucleotides Paired ends 4. Data Analysis Assembly of overlapping fragments A C Sequencing by synthesis G T Visualization of nucleotides being added to clusters

Genomic DNA and beyond

Pac. Bio SMRT – long read example Considerations • Sequencing libraries • No amplification • Sequence by synthesis • Long reads • Data analysis • Sequence assembly Vocabulary • Phospholinked label • Single molecule sequencing • SMRT cell • ZMW

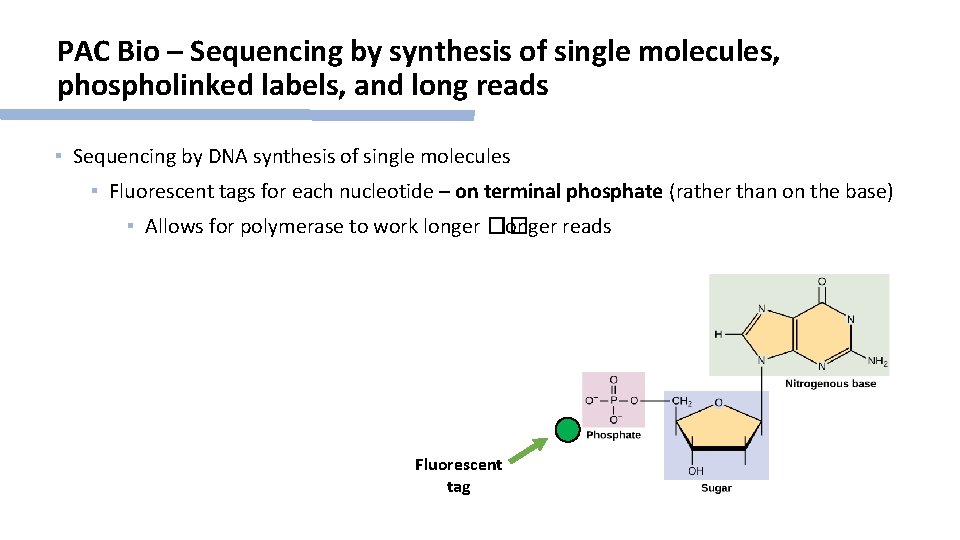
PAC Bio – Sequencing by synthesis of single molecules, phospholinked labels, and long reads ▪ Sequencing by DNA synthesis of single molecules ▪ Fluorescent tags for each nucleotide – on terminal phosphate (rather than on the base) ▪ Allows for polymerase to work longer �� longer reads Fluorescent tag

PAC Bio – SMRT ZMWs ▪ Single Molecule Real Time (SMRT) cells containing Zero Mode Waveguides (ZMWs) ▪ Sequential bursts of light corresponding to incorporation of nucleotides ▪ Advantages: ▪ No amplification of template required ▪ Minimizes base call errors that are created by fragment clusters SMRT cell 1 million ZMWs ZMW (70 nm wide) = 1 DNA template + 1 polymerase

PAC Bio – Sample preparation and libraries

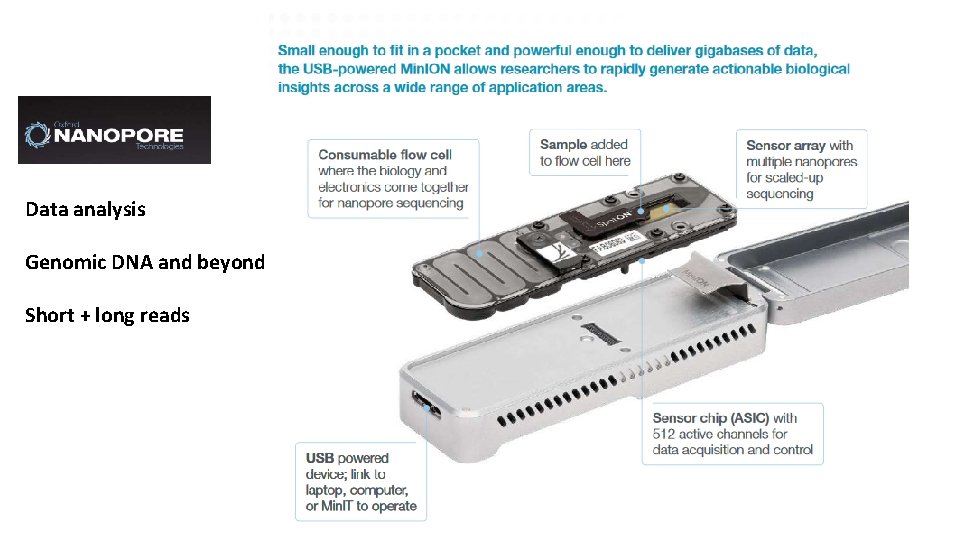
Data analysis Genomic DNA and beyond Short + long reads

Nanopore Sequencing Considerations • DNA • Sequencing libraries • Optional amplification • Single molecule sequencing • Long reads • Data analysis • Sequence assembly Vocabulary • Nanopore • Tether • Ionic current disruption

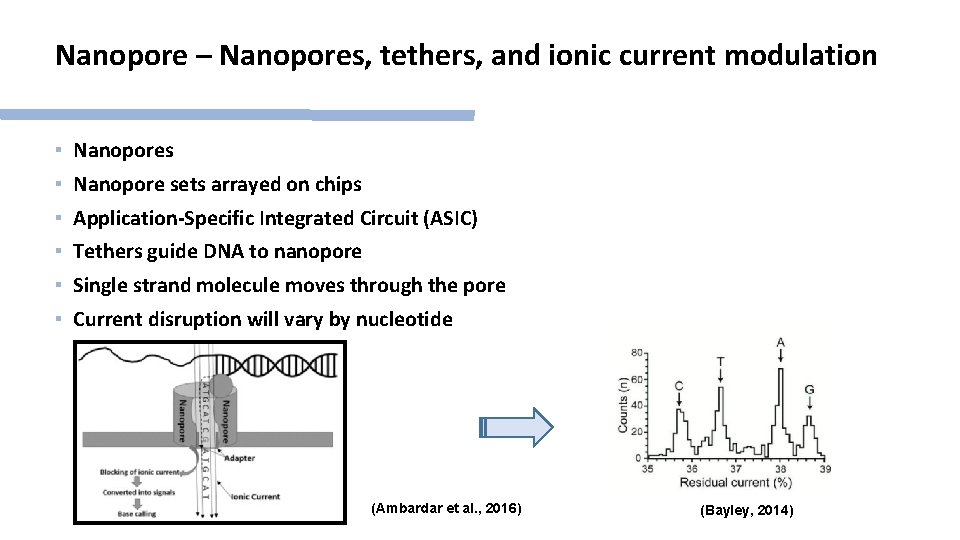
Nanopore – Nanopores, tethers, and ionic current modulation ▪ Nanopores ▪ Nanopore sets arrayed on chips ▪ Application-Specific Integrated Circuit (ASIC) ▪ Tethers guide DNA to nanopore ▪ Single strand molecule moves through the pore ▪ Current disruption will vary by nucleotide (Ambardar et al. , 2016) (Bayley, 2014)

Nanopore – Sample preparation and libraries High quality DNA essential for long reads DNA fragmentation Adapter ligation PCR amplification (optional) Attachment of tether protein to 5’ end of DNA

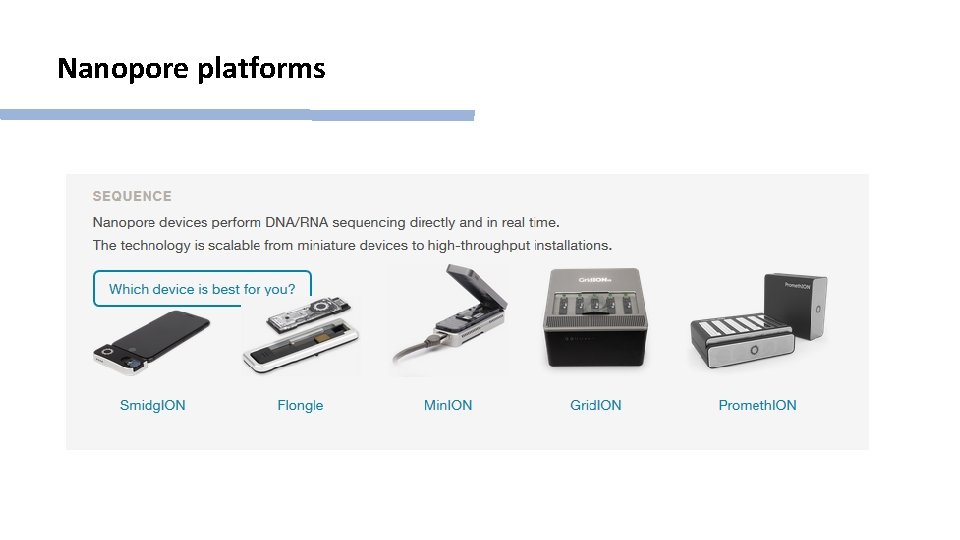
Nanopore platforms

Data analysis Genomic DNA and beyond Short + long reads

Features Illumina PAC Bio Nanopore Principle Sequence by synthesis Ionic current modulation 100’s of bp 10’s of kb 100’s of kb Fluorescence tagged nucleotides? Yes. Base is tagged. Yes. Terminal phosphate is tagged. No Number of copies of each fragment Multiple Single No No Yes (some versions) Read length Portable?

By now, you should be able to… ▪ For the Illumina short read technique, explain the following: ▪ The sequencing principle. ▪ Why cluster generation is needed, and the role of adapters in cluster generation. ▪ Why short reads can be a challenge for assembly but at the same time be useful for accurate genome sequencing. ▪ For the PAC Bio SMRT long read technique, explain the following: ▪ The sequencing principle. ▪ The advantages of having the fluorescent tag in the phosphate terminal of the nucleotide, instead of on the base. ▪ Why single molecule long reads can be an advantage, but may be best when combined with short reads. ▪ For the Nanopore technique, explain the following: ▪ The sequencing principle. ▪ List some advantages of this technology, compared to the Illumina and PAC Bio. ▪ If Nanopore is so great, why other options are still available.