Computational and experimental analysis of DNA shuffling Supporting







- Slides: 18

Computational and experimental analysis of DNA shuffling : Supporting text N. Maheshri and D. V. Schaffer PNAS, vol. 100, no. 6, 3071 -3076 Summarized by Shin, Soo-Yong

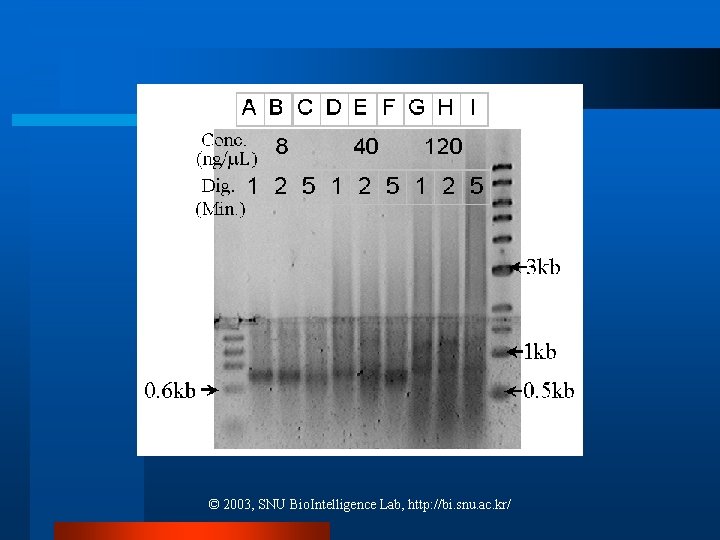
Length Distribution Comparison l Experimental and simulation data cannot be directly compared due to an agarose gel. ¨ Fragments migrate due to both the external electric field and molecular diffusion ¨ The intensity of fragments is related to both their concentration and size l Solution: Gaussian distribution ¨ Fitting to each band by mean and standard deviation © 2003, SNU Bio. Intelligence Lab, http: //bi. snu. ac. kr/

Length Distribution Comparison l Gaussian transformation function can be limited in accuracy for large fragments (> 2 kb) ¨ because of retarded mobility l Gaussian transformation function can be limited for small fragments (< 200 bp) because of difficulties with detection ¨ Small fragments bind less dye and diffusion significantly disperses them © 2003, SNU Bio. Intelligence Lab, http: //bi. snu. ac. kr/

Length Distribution Comparison l Detection can be improved by loading a greater amount of DNA ¨ Overloading a gel results in retarded mobility of the fragments: DNA steaking © 2003, SNU Bio. Intelligence Lab, http: //bi. snu. ac. kr/

© 2003, SNU Bio. Intelligence Lab, http: //bi. snu. ac. kr/

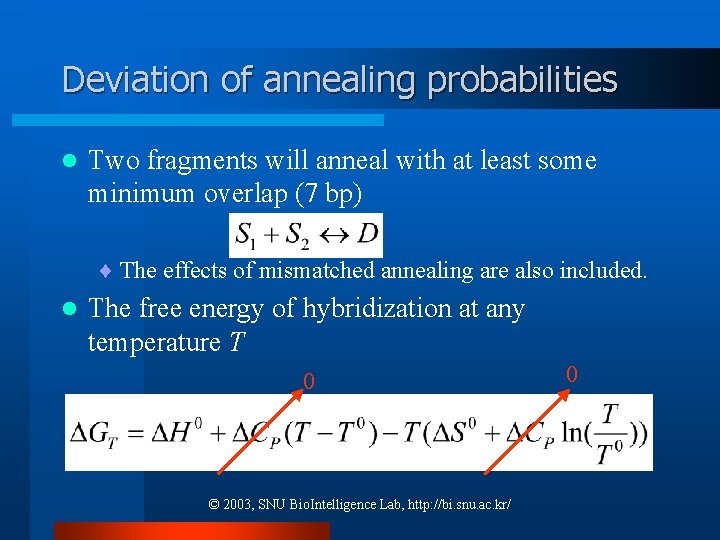
Deviation of annealing probabilities l Two fragments will anneal with at least some minimum overlap (7 bp) ¨ The effects of mismatched annealing are also included. l The free energy of hybridization at any temperature T 0 © 2003, SNU Bio. Intelligence Lab, http: //bi. snu. ac. kr/ 0

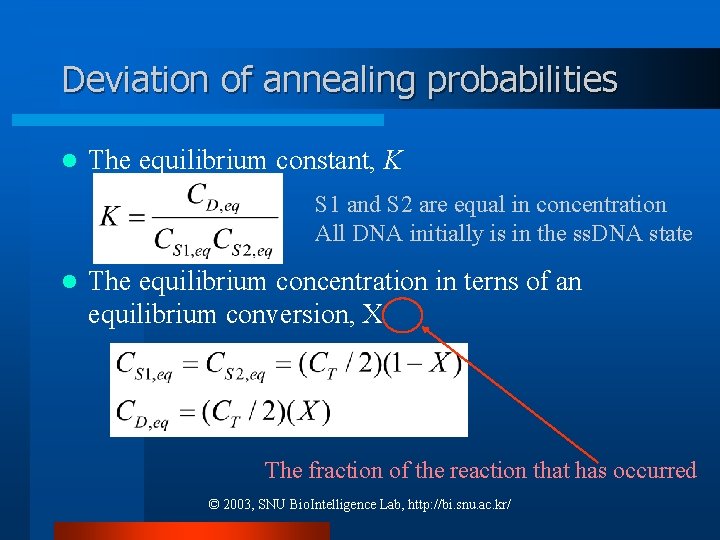
Deviation of annealing probabilities l The equilibrium constant, K S 1 and S 2 are equal in concentration All DNA initially is in the ss. DNA state l The equilibrium concentration in terns of an equilibrium conversion, X The fraction of the reaction that has occurred © 2003, SNU Bio. Intelligence Lab, http: //bi. snu. ac. kr/

Deviation of annealing probabilities X = 0 implies no reactants have converted to products. l X = 1 implies all reactants have converted to products. l l The equilibrium conversion Two distinct sequence : 4 Self-complementary : 1 © 2003, SNU Bio. Intelligence Lab, http: //bi. snu. ac. kr/

Deviation of annealing probabilities l Total number of ways, N, that S 1 and S 2 can anneal, given fragment lengths l 1 and l 2, and a minimum overlap v ¨ No gap or bulges ¨ Two ss. DNAs stretched out ¨ With dangling ends © 2003, SNU Bio. Intelligence Lab, http: //bi. snu. ac. kr/

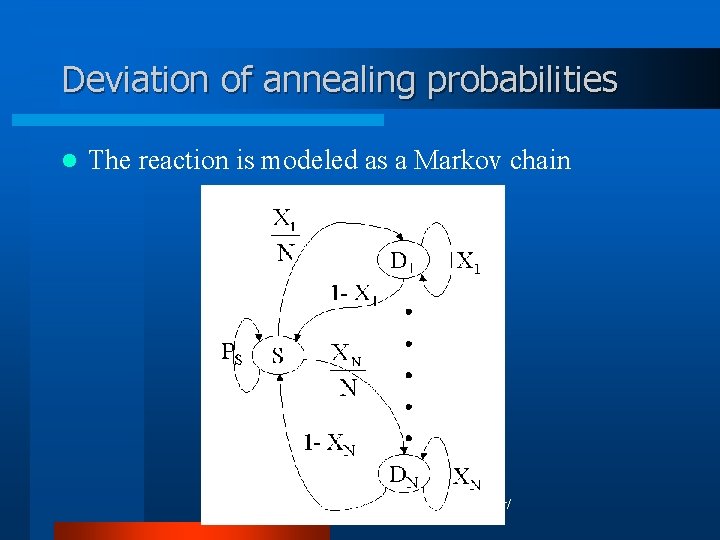
Deviation of annealing probabilities l The reaction is modeled as a Markov chain © 2003, SNU Bio. Intelligence Lab, http: //bi. snu. ac. kr/

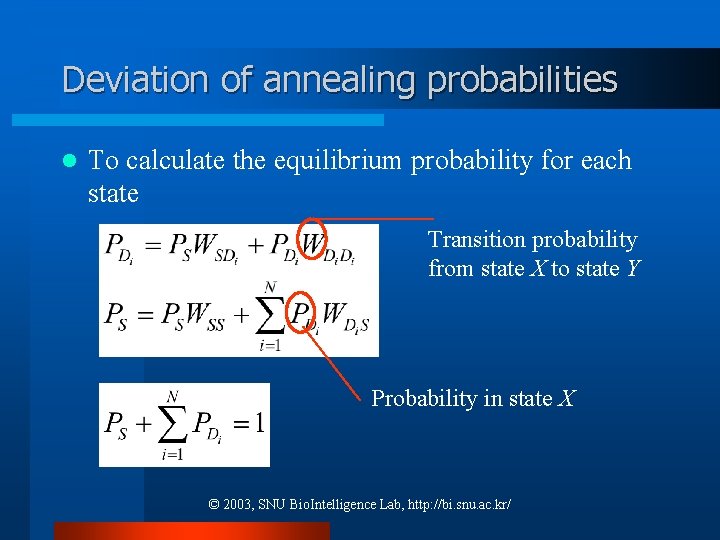
Deviation of annealing probabilities l To calculate the equilibrium probability for each state Transition probability from state X to state Y Probability in state X © 2003, SNU Bio. Intelligence Lab, http: //bi. snu. ac. kr/

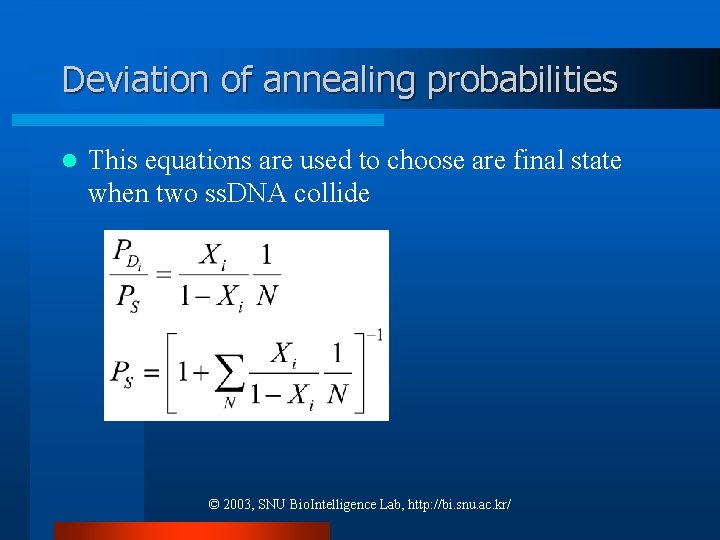
Deviation of annealing probabilities l This equations are used to choose are final state when two ss. DNA collide © 2003, SNU Bio. Intelligence Lab, http: //bi. snu. ac. kr/

Effects of concentration and temperature on hybridization stringency Changing temperature alters annealing probabilities in a strongly sequence-dependent fashion l Changes in concentration alter them in a more nonspecific manner l © 2003, SNU Bio. Intelligence Lab, http: //bi. snu. ac. kr/

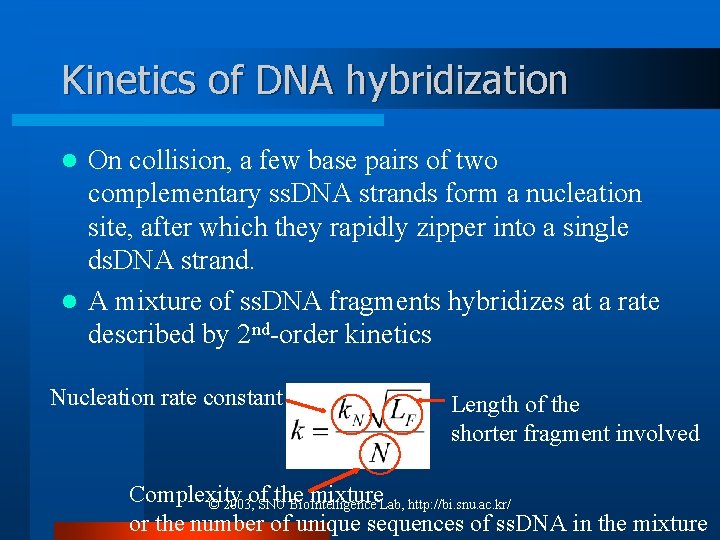
Kinetics of DNA hybridization On collision, a few base pairs of two complementary ss. DNA strands form a nucleation site, after which they rapidly zipper into a single ds. DNA strand. l A mixture of ss. DNA fragments hybridizes at a rate described by 2 nd-order kinetics l Nucleation rate constant Length of the shorter fragment involved Complexity of the mixture © 2003, SNU Bio. Intelligence Lab, http: //bi. snu. ac. kr/ or the number of unique sequences of ss. DNA in the mixture

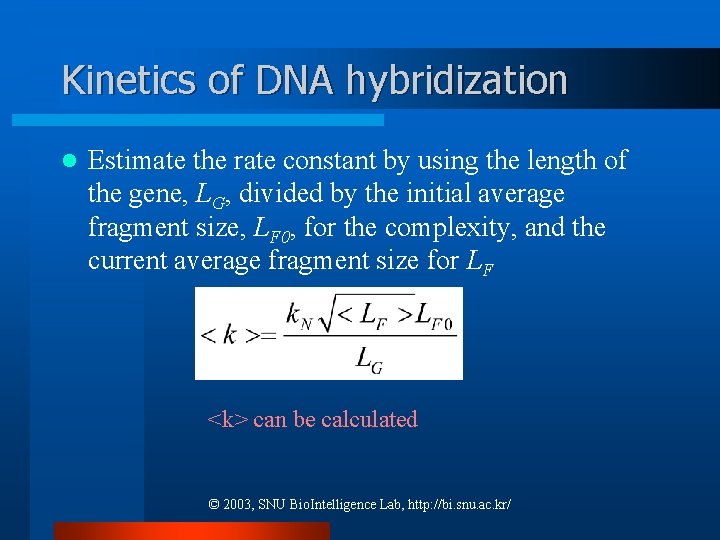
Kinetics of DNA hybridization l Estimate the rate constant by using the length of the gene, LG, divided by the initial average fragment size, LF 0, for the complexity, and the current average fragment size for LF <k> can be calculated © 2003, SNU Bio. Intelligence Lab, http: //bi. snu. ac. kr/

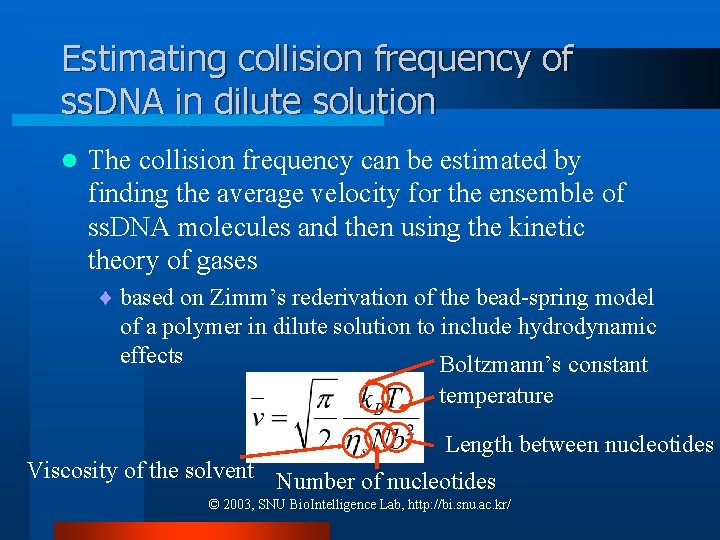
Estimating collision frequency of ss. DNA in dilute solution l The collision frequency can be estimated by finding the average velocity for the ensemble of ss. DNA molecules and then using the kinetic theory of gases ¨ based on Zimm’s rederivation of the bead-spring model of a polymer in dilute solution to include hydrodynamic effects Boltzmann’s constant temperature Length between nucleotides Viscosity of the solvent Number of nucleotides © 2003, SNU Bio. Intelligence Lab, http: //bi. snu. ac. kr/

Estimating collision frequency of ss. DNA in dilute solution l Average velocity of each particle and the mean free path, λ, between the particles, assuming the gas particle are ideal (neither interact nor take up volume) and spherical, Average radius of each particle Mean free path Number of particles per unit volume © 2003, SNU Bio. Intelligence Lab, http: //bi. snu. ac. kr/

Estimating collision frequency of ss. DNA in dilute solution l We can estimate the radius of a DNA molecule by its radius of gyration l For an aqueous solution of ss. DNA molecules at 0. 3 μM (corresponding to approximately 1 μg of digested fragments with an average fragment size of 100 bp in a 50 μl volume) at a temperature of 60°C ¨ an estimated collision frequency of ≈25 collisions per second per molecule. © 2003, SNU Bio. Intelligence Lab, http: //bi. snu. ac. kr/