DNA Shuffling the In Vitro Molecular Evolution Technique







- Slides: 16

DNA Shuffling, the In Vitro Molecular Evolution Technique, and Its Use in the Initial Pool Generation to Solve 26 -Cities TSP Ji Youn Lee School of Chemical Engineering Seoul National University

References • W. P. C. Stemmer, DNA shuffling by random fragmentation and reassembly In vitro recombination for molecular evolution Proc. Natl. Acad. Sci. USA (1994) 91 pp. 10747~10751 • Fengzhu Sun, Modeling DNA shuffling

DNA Shuffling? !

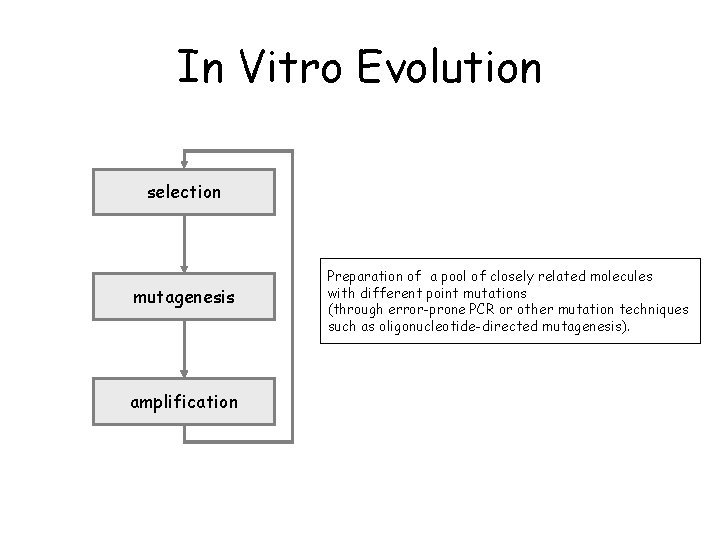
In Vitro Evolution selection mutagenesis amplification Preparation of a pool of closely related molecules with different point mutations (through error-prone PCR or other mutation techniques such as oligonucleotide-directed mutagenesis).

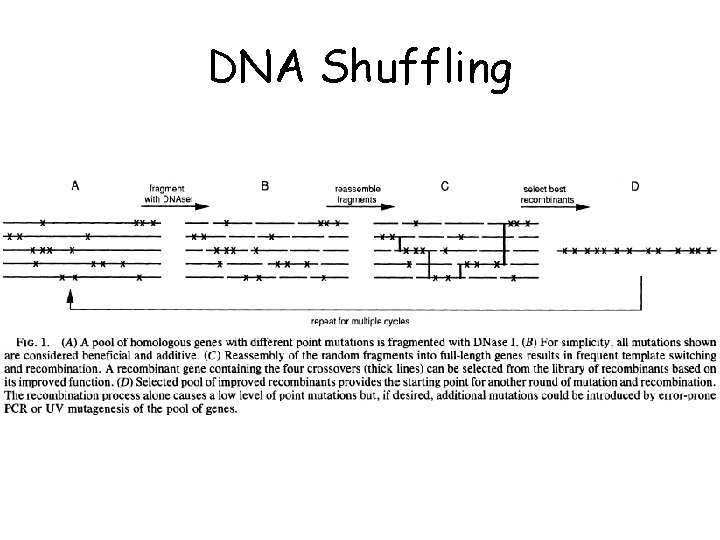
DNA Shuffling

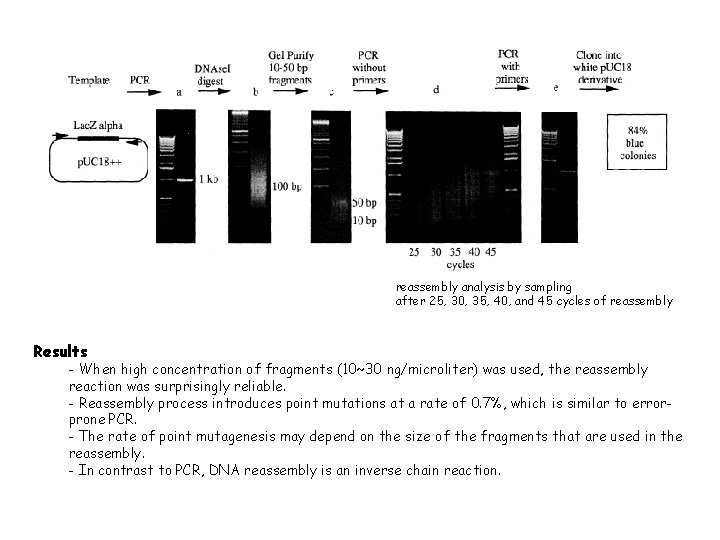
Substrate preparation DNase I digestion Sampling of fragments of lengths within a certain range 1 kb ds. DNA PCR products derived from p. UC 18 (reomoval of free primers) 2~4 ㎍ of the DNA substrate + 0. 0015 unit of DNase I per ㎕ in 100 ㎕ of 50 m. M Tris-HCl, p. H 7. 4, 1 m. M Mg. Cls for 10~20 min at RT Fragments of 10~50 bp were purified from 2% low meltin point agarose gels PCR without added primers 10~30 ng/㎕ of purified fragments 94℃ for 1 min (94℃ for 0. 5 min, 50~55 ℃ for 0. 5 min and 72℃ for 0. 5 min) 72℃ for 5 min PCR with primers 1: 40 dilution of the primerless PCR product into PCR mixture with 0. 8 m. M each primer and ~15 additional cycles And… a single product of the correct size is typically obtained Cloning and analysis

reassembly analysis by sampling after 25, 30, 35, 40, and 45 cycles of reassembly Results - When high concentration of fragments (10~30 ng/microliter) was used, the reassembly reaction was surprisingly reliable. - Reassembly process introduces point mutations at a rate of 0. 7%, which is similar to errorprone PCR. - The rate of point mutagenesis may depend on the size of the fragments that are used in the reassembly. - In contrast to PCR, DNA reassembly is an inverse chain reaction.

Its Application to the Initial Pool Generation

Advantages • More economic! – – No need of phosphorylation No need of ligase (terrible labour of course…) d. NTPs are much cheaper than oligomers We can use the saved money for the study of bead separation • More reliable! – No need of hybridization/ligation step – Lower concentration of the initial olgomers is tolerable? ! – We believe the potential of PCR • Originality? !

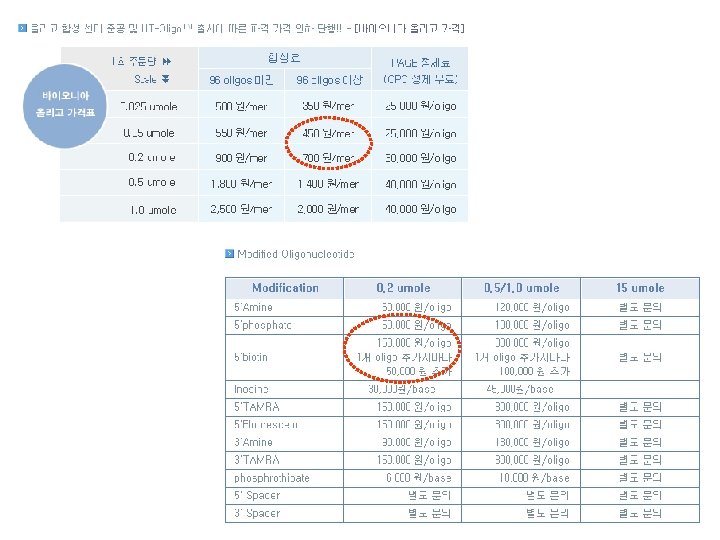
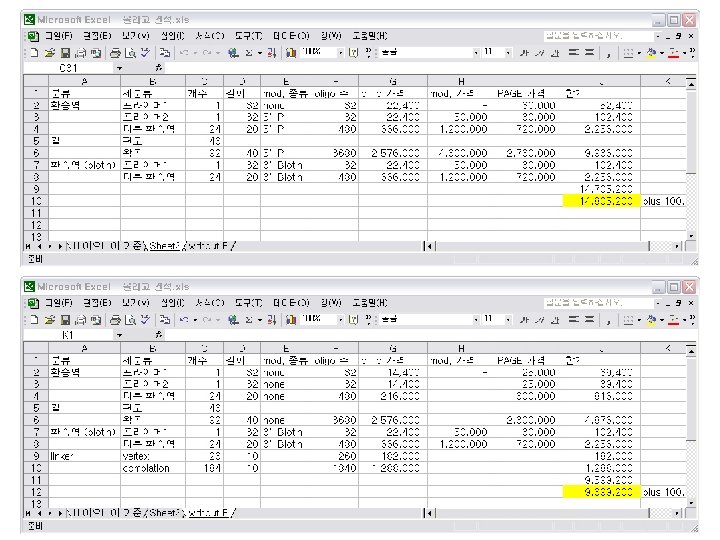
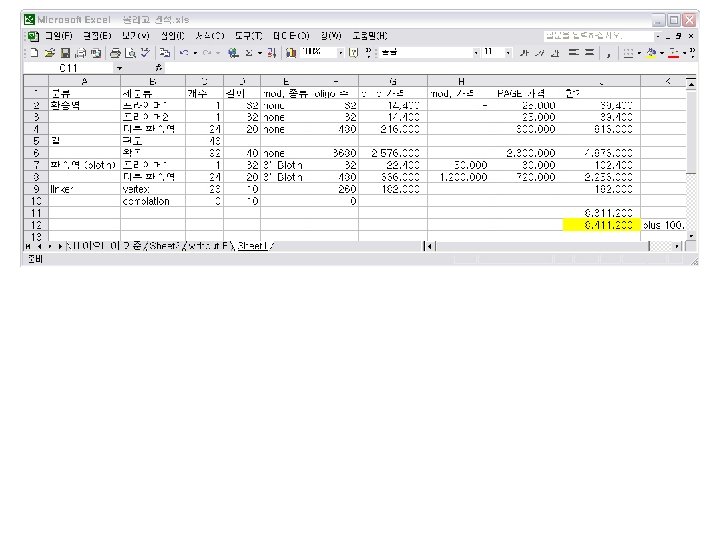
An Estimate of Oligomer Cost




Disadvantages • I have no experience! • I have no advisor! • Is it possible in the real world?

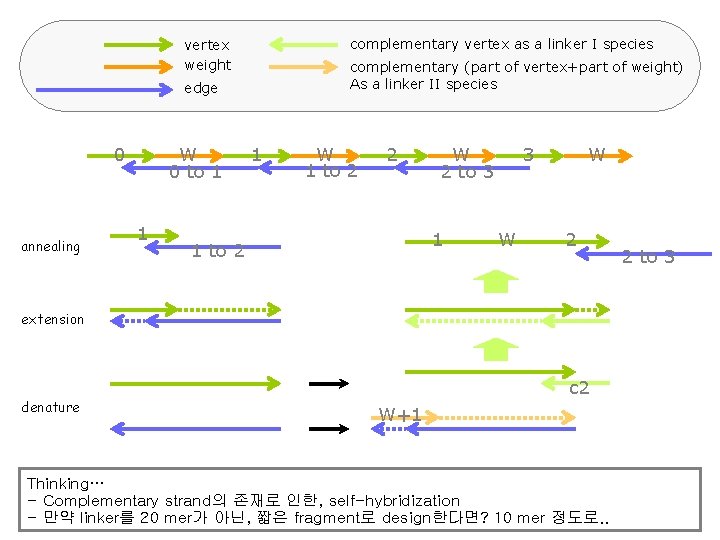
How It Works?

complementary vertex as a linker I species vertex weight complementary (part of vertex+part of weight) As a linker II species edge 0 annealing W 0 to 1 1 1 W 1 to 2 2 W 2 to 3 1 1 to 2 3 W W 2 extension denature c 2 W+1 Thinking… - Complementary strand의 존재로 인한, self-hybridization - 만약 linker를 20 mer가 아닌, 짧은 fragment로 design한다면? 10 mer 정도로. . 2 to 3