Cancer Res 1982 Mar 423 1088 93 Cloning























































- Slides: 63





Cancer Res. 1982 Mar; 42(3): 1088 -93. Cloning and screening of sequences expressed in a mouse colon tumor. Augenlicht LH, Kobrin D. Memorial Sloan-Kettering Cancer Center, Walker Laboratory, Rye, New York 10580 Polyadenylic acid-containing cytoplasmic RNA was isolated from a dimethylhydrazineinduced mouse transplantable colon carcinoma. Double-stranded complementary DNA synthesized using these molecules was inserted into the Hind. III site of p. BR 322 using Hind. III linkers, and the recombinant molecules were cloned in Escherichia coli. Clones were screened with labeled complementary DNA synthesized from the polyadenylic acidcontaining cytoplasmic RNA of the tumor or normal mouse colon, liver, or kidney. Of 378 clones screened with the normal colon and tumor probes, seven showed major increases in abundance in the tumor tissue as compared to normal, and one showed a major decrease. Twenty-five other sequences were found which showed smaller increases and 22 showed smaller decreases. Of 373 clones screened with tumor, colon, liver, and kidney probes, 79% exhibited little evidence of tissue specificity in expression. The data for those sequences which do show tissue-specific regulation of expression suggest that the tumor continues to express sequences characteristic of the colon but also shows a loss, or decrease, in other gene products, most of which (19 of 25) are characteristic of the colon. Finally, nine of 10 sequences which increase from low abundance in the normal colon to high abundance in the tumor are found at a moderate to high level in the liver and kidney. On the other hand, 23 of 36 sequences which show more modest increases in the tumor as compared to normal are scarce or absent in the liver and kidney. PMID: 7059971 [Pub. Med - indexed for MEDLINE]

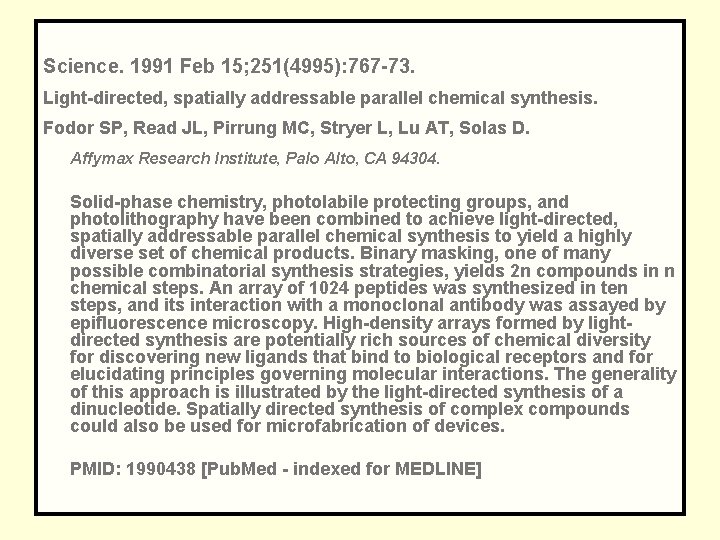
Science. 1991 Feb 15; 251(4995): 767 -73. Light-directed, spatially addressable parallel chemical synthesis. Fodor SP, Read JL, Pirrung MC, Stryer L, Lu AT, Solas D. Affymax Research Institute, Palo Alto, CA 94304. Solid-phase chemistry, photolabile protecting groups, and photolithography have been combined to achieve light-directed, spatially addressable parallel chemical synthesis to yield a highly diverse set of chemical products. Binary masking, one of many possible combinatorial synthesis strategies, yields 2 n compounds in n chemical steps. An array of 1024 peptides was synthesized in ten steps, and its interaction with a monoclonal antibody was assayed by epifluorescence microscopy. High-density arrays formed by lightdirected synthesis are potentially rich sources of chemical diversity for discovering new ligands that bind to biological receptors and for elucidating principles governing molecular interactions. The generality of this approach is illustrated by the light-directed synthesis of a dinucleotide. Spatially directed synthesis of complex compounds could also be used for microfabrication of devices. PMID: 1990438 [Pub. Med - indexed for MEDLINE]










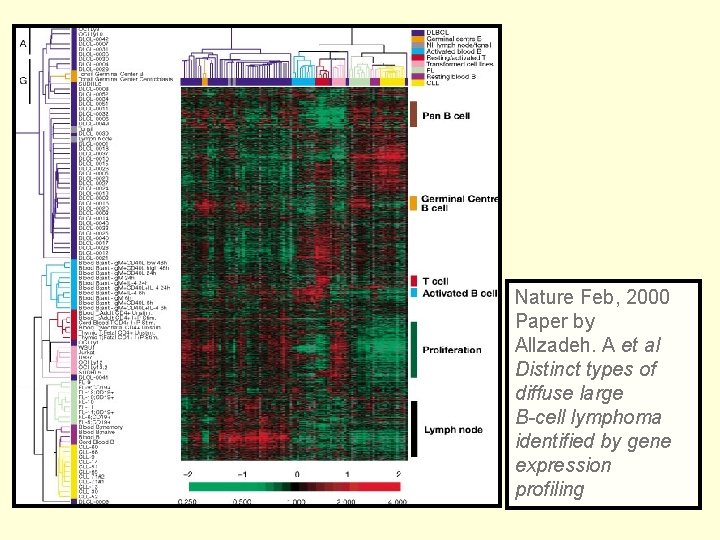
Nature Feb, 2000 Paper by Allzadeh. A et al Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling





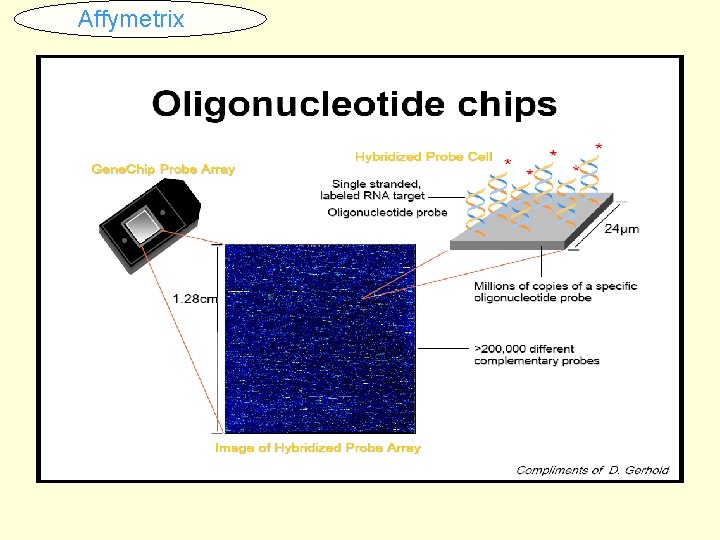
Affymetrix










Affymetrix


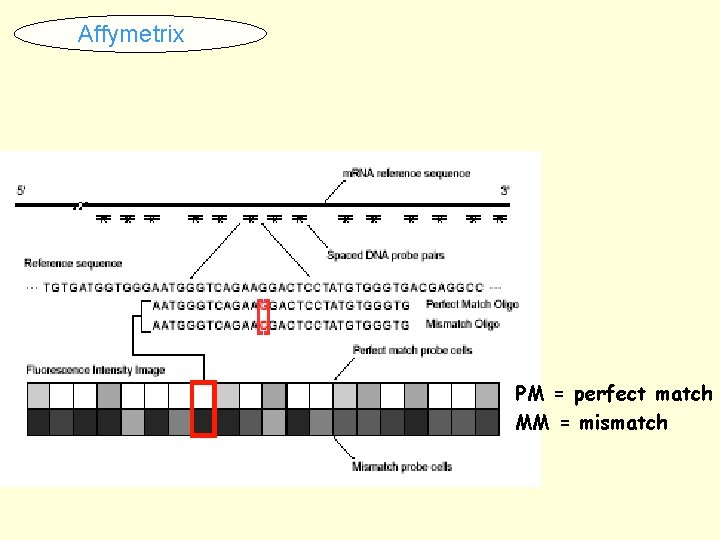
Affymetrix PM = perfect match MM = mismatch








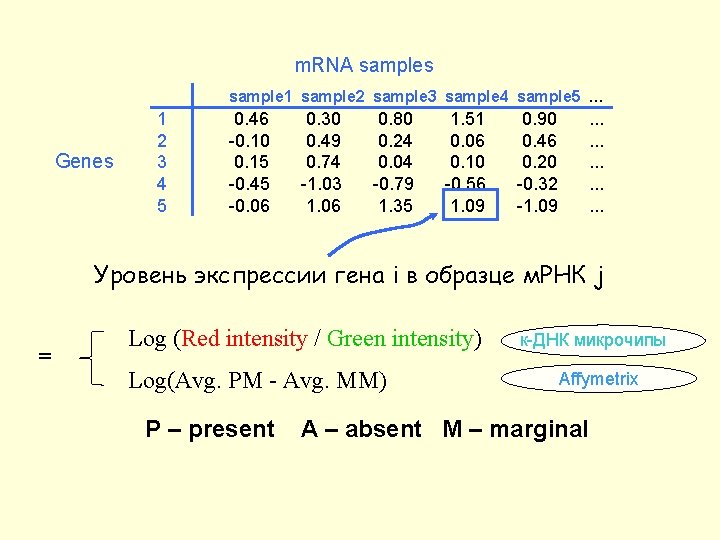
m. RNA samples sample 1 sample 2 sample 3 sample 4 sample 5 … Genes 1 2 3 4 5 0. 46 -0. 10 0. 15 -0. 45 -0. 06 0. 30 0. 49 0. 74 -1. 03 1. 06 0. 80 0. 24 0. 04 -0. 79 1. 35 1. 51 0. 06 0. 10 -0. 56 1. 09 0. 90 0. 46 0. 20 -0. 32 -1. 09 . . . . Уровень экспрессии гена i в образце м. РНК j = Log (Red intensity / Green intensity) Log(Avg. PM - Avg. MM) P – present к-ДНК микрочипы Affymetrix A – absent M – marginal
















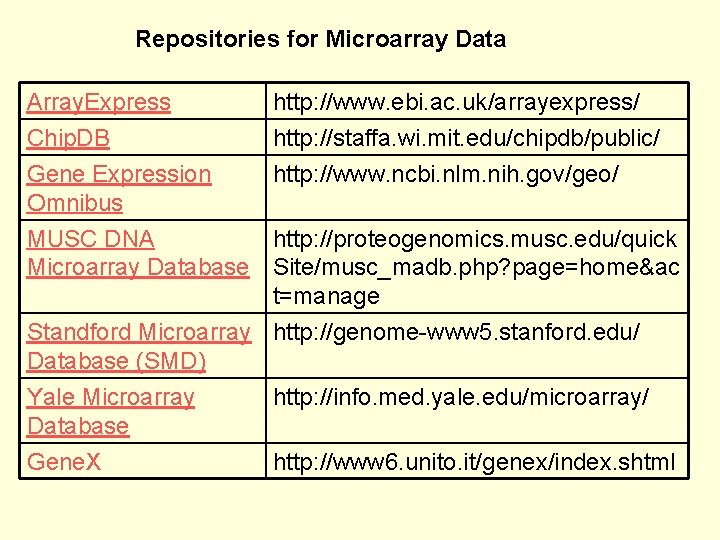
Repositories for Microarray Data Array. Express Chip. DB Gene Expression Omnibus http: //www. ebi. ac. uk/arrayexpress/ http: //staffa. wi. mit. edu/chipdb/public/ http: //www. ncbi. nlm. nih. gov/geo/ MUSC DNA http: //proteogenomics. musc. edu/quick Microarray Database Site/musc_madb. php? page=home&ac t=manage Standford Microarray http: //genome-www 5. stanford. edu/ Database (SMD) Yale Microarray http: //info. med. yale. edu/microarray/ Database Gene. X http: //www 6. unito. it/genex/index. shtml

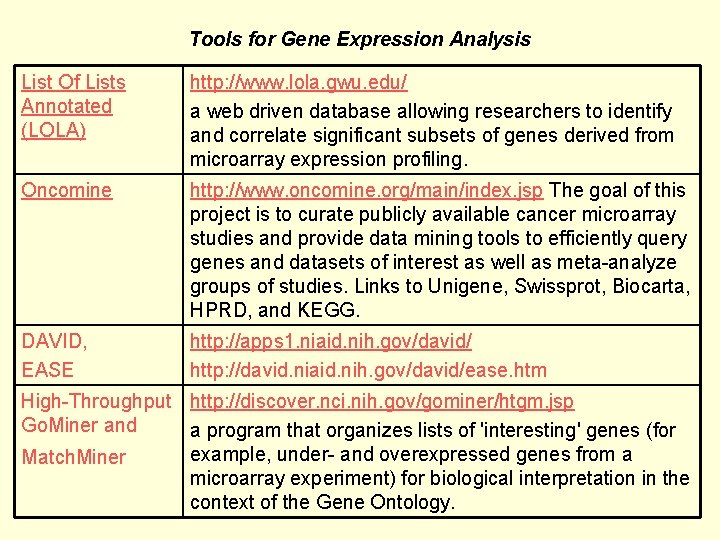
Tools for Gene Expression Analysis List Of Lists Annotated (LOLA) http: //www. lola. gwu. edu/ a web driven database allowing researchers to identify and correlate significant subsets of genes derived from microarray expression profiling. Oncomine http: //www. oncomine. org/main/index. jsp The goal of this project is to curate publicly available cancer microarray studies and provide data mining tools to efficiently query genes and datasets of interest as well as meta-analyze groups of studies. Links to Unigene, Swissprot, Biocarta, HPRD, and KEGG. DAVID, EASE http: //apps 1. niaid. nih. gov/david/ http: //david. niaid. nih. gov/david/ease. htm High-Throughput http: //discover. nci. nih. gov/gominer/htgm. jsp Go. Miner and a program that organizes lists of 'interesting' genes (for example, under- and overexpressed genes from a Match. Miner microarray experiment) for biological interpretation in the context of the Gene Ontology.





 Pengertian inbreng
Pengertian inbreng Res rei declinazione
Res rei declinazione Astronomo vermeer
Astronomo vermeer Secretaria de energia 1102
Secretaria de energia 1102 Identity cloning and concealment
Identity cloning and concealment Blunt end ligation
Blunt end ligation Advantages of cloning
Advantages of cloning Clock gate cloning
Clock gate cloning Process of cloning
Process of cloning Suspension cloning
Suspension cloning Therpeutic cloning
Therpeutic cloning Advantages and disadvantages of cloning
Advantages and disadvantages of cloning Cloning in nature
Cloning in nature Gene cloning
Gene cloning Reproductive cloning
Reproductive cloning Positional cloning
Positional cloning What is cloning
What is cloning M13 phage vector
M13 phage vector Genez molecular clones
Genez molecular clones Genetic engineering steps
Genetic engineering steps Cloning and sequencing explorer series
Cloning and sequencing explorer series Clonaid
Clonaid Gateway technology with clonase ii
Gateway technology with clonase ii Helpx
Helpx Plasmid
Plasmid Cloning vectors
Cloning vectors Cloning pros and cons
Cloning pros and cons Inverse pcr
Inverse pcr Gsm cloning
Gsm cloning History of cloning
History of cloning Nick translation
Nick translation Advantages and disadvantages of cloning
Advantages and disadvantages of cloning Elsa gunter
Elsa gunter 8774237243
8774237243 Ie 423
Ie 423 Cos 423
Cos 423 Cs 423
Cs 423 Ece 423
Ece 423 Brb 423
Brb 423 Princeton data structures and algorithms
Princeton data structures and algorithms Cse 423
Cse 423 Uiuc cs 423
Uiuc cs 423 Cont 423
Cont 423 Me 423
Me 423 Cos423
Cos423 Cse 423
Cse 423 Ai 423
Ai 423 Rlc
Rlc Me 423
Me 423 1982 loi de décentralisation
1982 loi de décentralisation 1998-1982
1998-1982 Howell 1982 conscious competence model
Howell 1982 conscious competence model Campeonato uruguayo 1982
Campeonato uruguayo 1982 1982-1938
1982-1938 Inang bayan english
Inang bayan english Maps approach to team building
Maps approach to team building Poltergeist 1982 screenplay by
Poltergeist 1982 screenplay by Normas iso dibujo tecnico
Normas iso dibujo tecnico Howell, 1982 conscious competence model
Howell, 1982 conscious competence model Permenaker no. 03/men/1982
Permenaker no. 03/men/1982 1982-1973
1982-1973 1982-1938
1982-1938 Tim crawford cpa
Tim crawford cpa Carron's 1982 model of cohesion
Carron's 1982 model of cohesion