Automatic QRS Complex Detection Algorithm Designed for a















- Slides: 19

Automatic QRS Complex Detection Algorithm Designed for a Novel Wearable, Wireless Electrocardiogram Recording Device - Presented By Sagar Naik ( 1703005 ) Shraddha Pradhan ( 1703007 )

2 ❑ Introduction ❑ Methodology Contents ❑ Results ❑ Conclusion ❑ References

3 Introduction

4 ▸Ambulatory ECG monitoring are numerous. ▸DELTA has developed the e. Patch. Need ▸ECG channels on the sternum. ▸Do not correspond to any standard HOLTER leads.

5 ▸ECG analysis is a robust, reliable and automatic QRS detection algorithm. QRS Detection ▸Optimized for the special e. Patch ECG signals. ▸Several different approaches. ▸Automatic QRS detection ▸Noise ▸Computational Complexity

6 To overcome some of the limitations, the proposed QRS detection algorithm can be applied in two different modes: Single-channel and Multi-channel mode Single Channel & Multichannel Mode ▸Multi-channel mode. ▸Single-channel mode. ▸ QRS detection is generally based on channel I of the MIT -BIH Arrhythmia Database (MITDB)

7 Methodology

8 ▸Monitoring of 11 different patients. ▸Two ECG channels recording with a sampling frequency of 500 Hz and a resolution of 13 bits. Data ▸ 30 minutes were extracted, one hour after beginning of the recording. ▸ All beats annotated as “normal”.

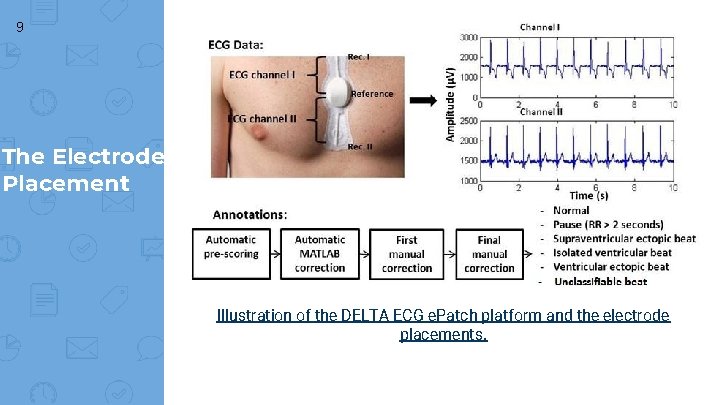
9 The Electrode Placement Illustration of the DELTA ECG e. Patch platform and the electrode placements.

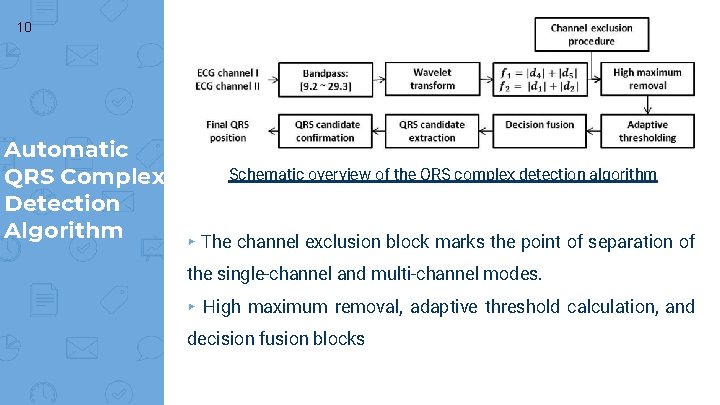
10 Automatic Schematic overview of the QRS complex detection algorithm QRS Complex Detection Algorithm ▸ The channel exclusion block marks the point of separation of the single-channel and multi-channel modes. ▸ High maximum removal, adaptive threshold calculation, and decision fusion blocks

11 ▸ Channel Exclusion Criteria. ▸Bandpass Filtering ▸Wavelet Transform Algorithm

12 ▸ Detection of QRS Candidates. ▸An adaptive threshold QRS Localization and Confirmation Block. ▸The location of the second zero-crossing is applied if more than one zero-crossing occurred in the bandpass filtered signal during this time interval.

13 Results

14 ▸ Beat detection accuracy ▸gross and the average statistics. ▸Using only channel I, only channel II (single-channel modes) and both channels (multi-channel mode).

15 Conclusion

16 ▸Achieves good performance. ▸Improved by implementation of more sophisticated channel exclusion criteria. ▸High detection sensitivity to abnormal beats.

17 References

18 ▸ F. Chiarugi, V. Sakkalis, D. Emmanouilidou, T. Krontiris, M. Varanini, and I. Tollis, Adaptive threshold QRS detector with best channel selection based on a noise rating system, Comput. Cardiol. , pp. 157160, 2007 ▸ H. Boqiang and W. Yuanyuan, Detecting QRS complexes of two -channel ECG signals by using combined wavelet entropy, 3 rd Inter-national Conference on Bioinformatics and Biomedical Engineering, vols. 1 -11, pp. 24392442, 2009.

19 THANKS!