Alu J Alu Jb Alu Jo Alu Sx
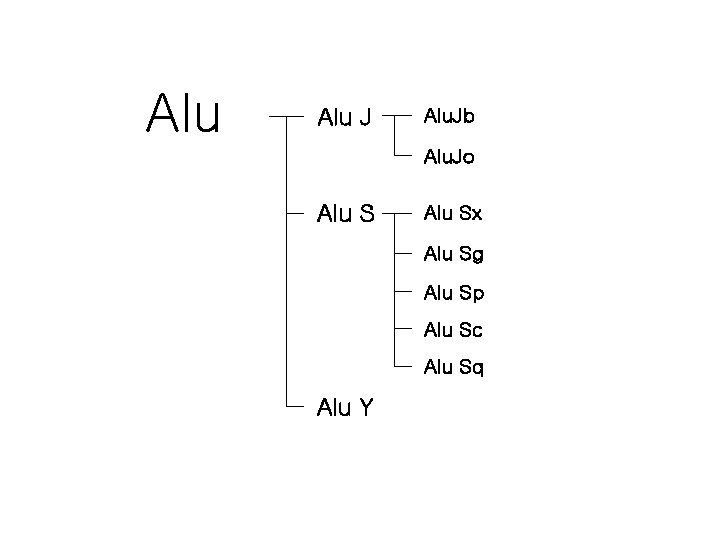
Alu J Alu. Jb Alu. Jo Alu Sx Alu Sg Alu Sp Alu Sc Alu Sq Alu Y
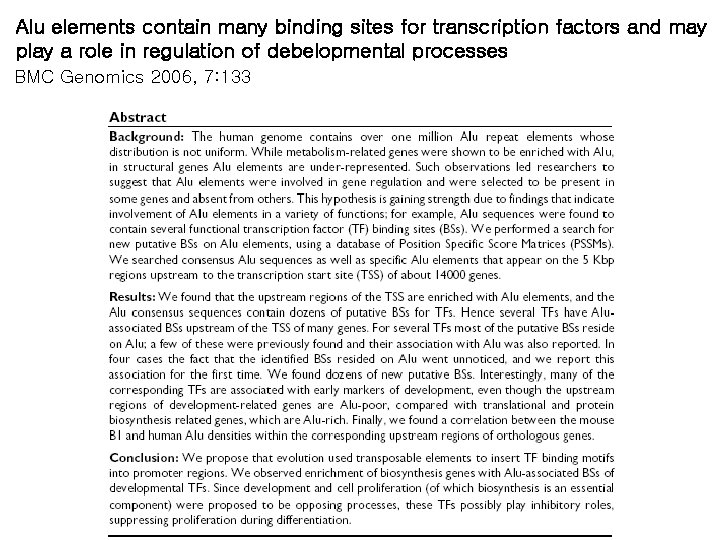
Alu elements contain many binding sites for transcription factors and may play a role in regulation of debelopmental processes BMC Genomics 2006, 7: 133

Feature of Alus Distribution of Alu was found to be highly correlated with local GC content and with the density of genes and with intron density Density of Alu is higher in intragenic(12. 5% of the nucleotides) than in intergenic regions(9. 6% of the nucleotides) Alu elements are negatively selected in imprinted regions of primate genomes Almost 20% of the genes contain transposable elements in the 3’UTR Housekeeping genes contain more Alu than tissue specific genes

The idea of Alu acting as a carrier of cis regularoty elements was suggested by Britten ; Evolutionary selection against change in many Alu repeat sequences interspersed through primate genomes(1994) Our findings show for the first time that TFs that are known to regulate the developmental processes may bind to Alu elements that are incorporated in the promoter regions of genes that need to be activated or suppressed during development






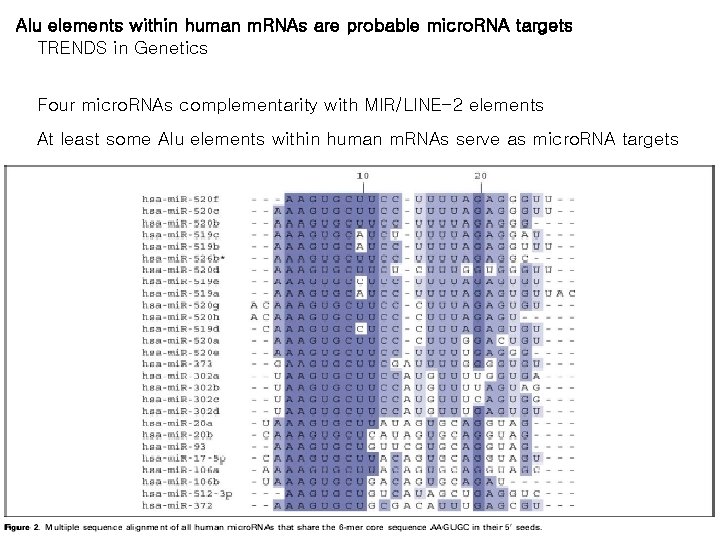
Alu elements within human m. RNAs are probable micro. RNA targets TRENDS in Genetics Four micro. RNAs complementarity with MIR/LINE-2 elements At least some Alu elements within human m. RNAs serve as micro. RNA targets

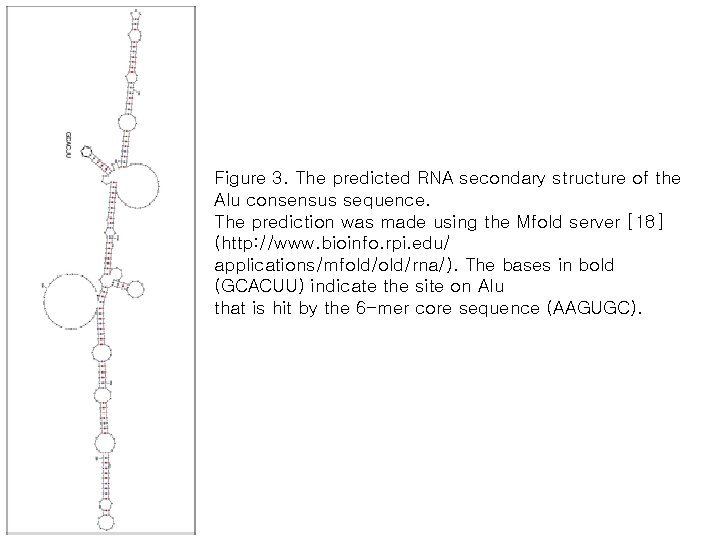
Figure 3. The predicted RNA secondary structure of the Alu consensus sequence. The prediction was made using the Mfold server [18] (http: //www. bioinfo. rpi. edu/ applications/mfold/rna/). The bases in bold (GCACUU) indicate the site on Alu that is hit by the 6 -mer core sequence (AAGUGC).

There is no biological reason why Alu or other transposable elements within m. RNAs should be regarded as unavailable for regulation by micro. RNAs Indeed m. RNAs that contain multiple Alu elements within their 3’UTRs should be regarded as particularly promising targets
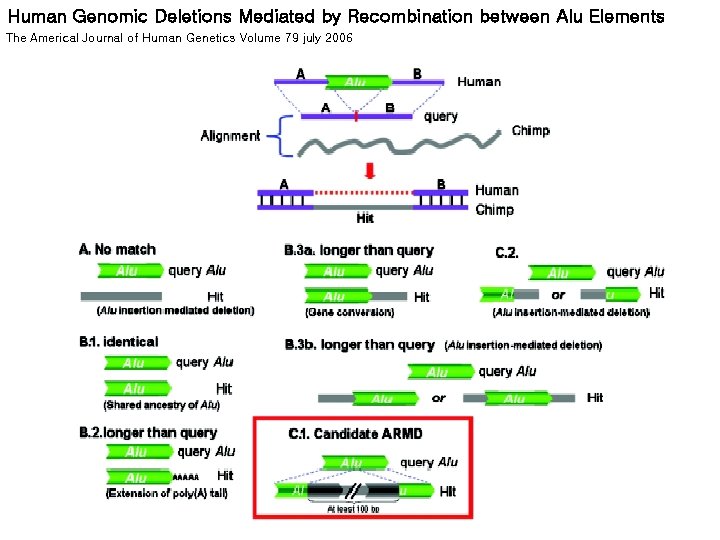
Human Genomic Deletions Mediated by Recombination between Alu Elements The Americal Journal of Human Genetics Volume 79 july 2006






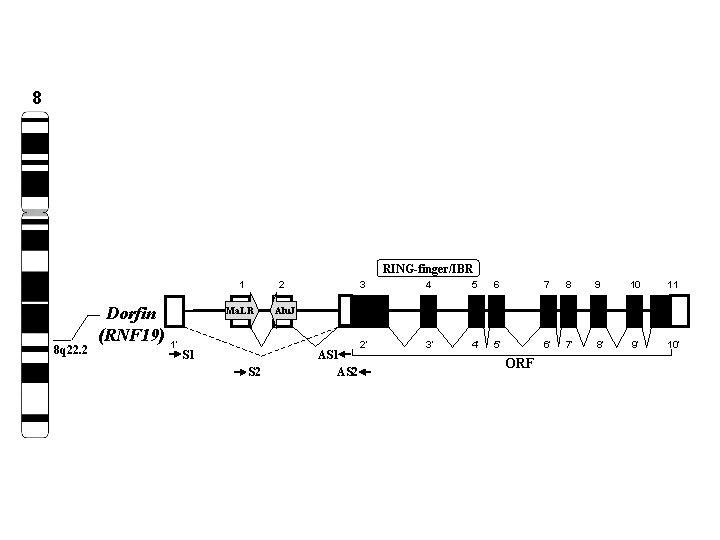
8 RING-finger/IBR 1 8 q 22. 2 Dorfin (RNF 19) 2 Ma. LR 1’ S 1 S 2 3 4 5 6 7 8 9 10 11 2’ 3’ 4’ 5’ 6’ 7’ 8’ 9’ 10’ Alu. J AS 1 AS 2 ORF
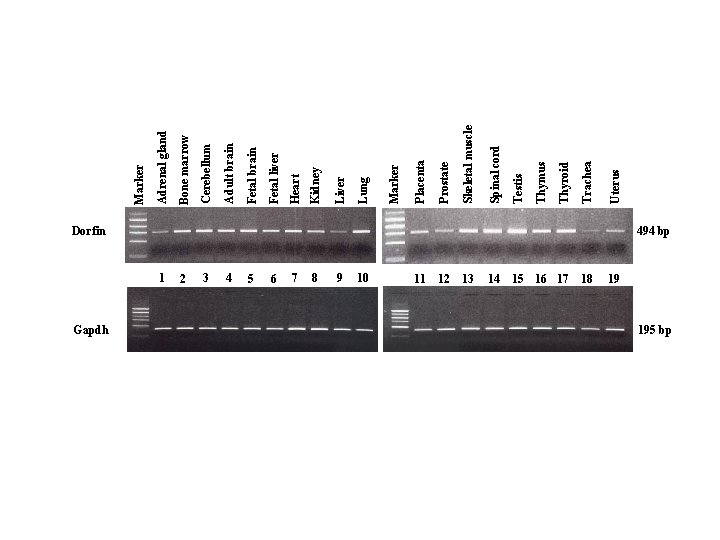
Dorfin Gapdh 494 bp 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 195 bp Uterus Trachea Thyroid Thymus Testis Spinal cord Skeletal muscle Prostate Placenta Marker Lung Liver Kidney Heart Fetal liver Fetal brain Adult brain Cerebellum Bone marrow Adrenal gland Marker
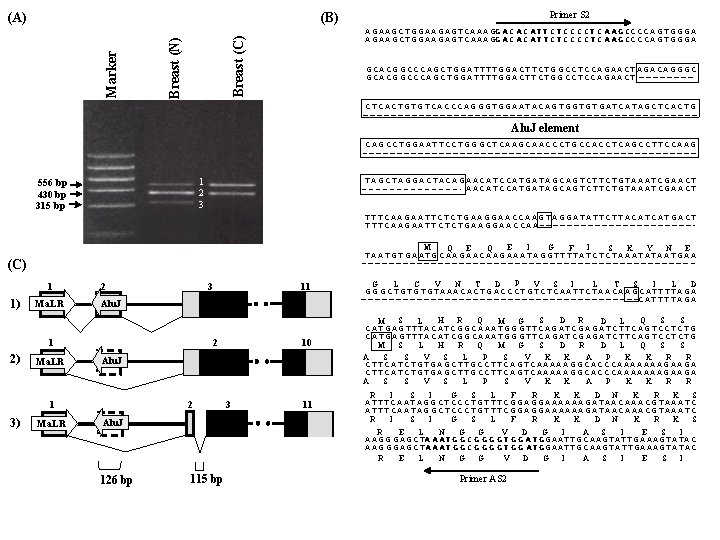
Primer S 2 (B) A G A A G C T G G A A G A G T C A A A GG A C A C A T T C T C C C C T C A A G CC C C A G T G G G A Breast (C) Breast (N) Marker (A) GCACGGCCCAGCTGGATTTTGGACTTCTGGCCTCCAGAACTAGACAGGGC G C A C G G C C C A G C T G G A T T G G A C T T C T G G C C T C C A G A A C TA G A C A G G G C CTCACTGTGTCACCCAGGGTGGAATACAGTGGTGTGATCATAGCTCACTG Alu. J element CAGCCTGGAATTCCTGGGCTCAAGCAACCCTGCCACCTCAGCCTTCCAAG 1 2 3 556 bp 430 bp 315 bp TAGCTAGGACTACAGAACATCCATGATAGCAGTCTTCTGTAAATCGAACT T A G C T A G G A C T A C A GA A C A T C C A T G A T A G C A G T C T G T A A A T C G A A C T TTTCAAGAATTCTCTGAAGGAACCAAGTAGGATATTCTTACATCATGACT T C A A G A A T T C T G A A G G A A C C A AG T A G G A T T C T T A C A T G A C T M (C) 1 1) Ma. LR 2 3 Ma. LR 2 Ma. LR 10 Alu. J 1 3) 11 Alu. J 1 2) E Q G I E Q I F K S Y E N TAATGTGAATGCAAGAACAAGAAATAGGTTTTATCTCTAAATATAATGAA G L V C T N P D V I S L S T I D L GGGCTGTGTGTAAACACTGACCCTGTCTCAATTCTAACAAGCATTTTAGA G G G C T G T G T A A A C T G A C C C T G T C A A T T C T A A C A A GC A T T A G A M S L H R Q M G S D R D L Q S S CATGAGTTTACATCGGCAAATGGGTTCAGATCGAGATCTTCAGTCCTCTG A S S V S L P S V K K A P K K R R CTTCATCTGTGAGCTTGCCTTCAGTCAAAAAGGCACCCAAAAAAAGAAGA 2 3 11 R I R Alu. J S I G S L F R K K D N K R K S ATTTCAATAGGCTCCCTGTTTCGGAGGAAAAAAGATAACAAACGTAAATC I S I G S L F R K K D N K R K S R E L N G G V D G I A S I E S I A A G G G A G C TA A A T G G C G G G G T G G A T GG A A T T G C A A G T A T T G A A A G T A T A C 126 bp 115 bp Primer AS 2
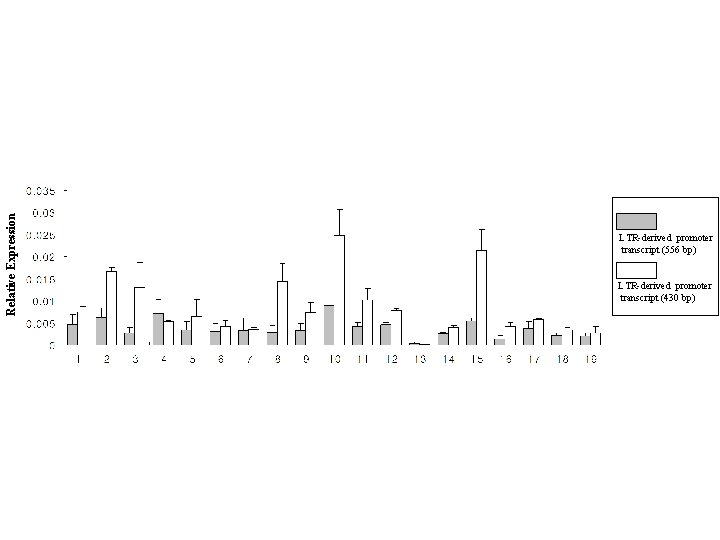
Relative Expression LTR-derived promoter transcript (556 bp) LTR-derived promoter transcript (430 bp)
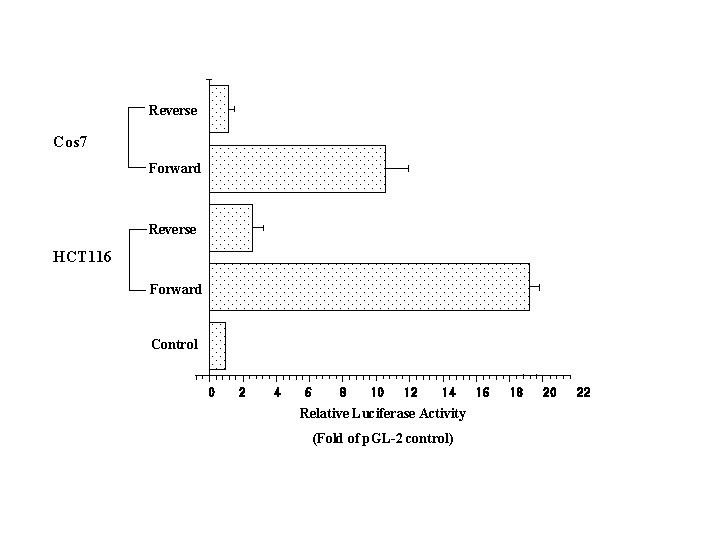
Reverse Cos 7 Forward Reverse HCT 116 Forward Control 0 2 4 6 8 10 12 14 Relative Luciferase Activity (Fold of p. GL-2 control) 16 18 20 22

GATA-1 CCCCACAAAA GATATGTTCA TGTCCTAATC CCCAGAATCT GCAAATGTTA TTTGGAAAAA GATA-2 GGGGTTTTGC AGATGTAATT AAGTTAAGAA TCTTGAGATA AGATCATCCT GGATTATCCA Nkx 2 -5 GGTAGCCTCA AAATCAAGTG ACAAGTGTCT TTGTAAGGGA CAAGTAGACC CATTACAGAG Whn ELF-1 AAGACGACGC GCAGAAAAGG AGGAAGCAGT GTGCTCATGG AGGCGGAGAT TGGAGTGATG +1 TRANSCRIPTION START SITE TAACCGCAAG CCGAGGAATG CTTATAGTCA CCAGAAGCTG GAAGAGTCAA AGGACACATT CTCCCCTCAA GCCCCAGTGG GAGCACGGCC CAGCTGGATT TTGGACTTCT GGCCTCCAGA ACTGTAAGAG AAATGTCCAT TGTCTTAAGC CAACCAGTTT GTGGTAGTTT GTTACAGCAG C C A G G A A A C T A .
100 77 42 100
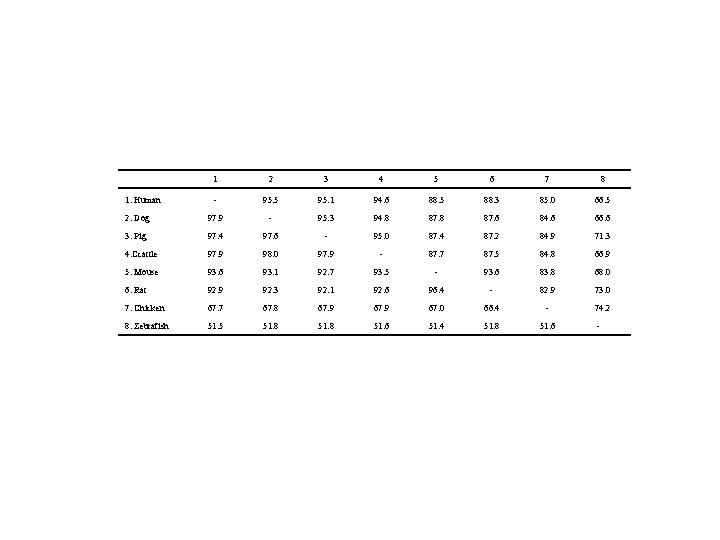
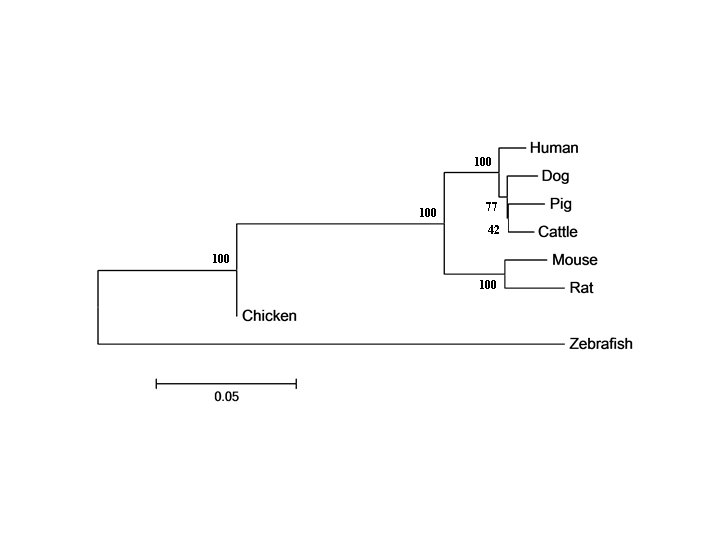
1 2 3 4 5 6 7 8 - 95. 5 95. 1 94. 6 88. 5 88. 3 85. 0 66. 5 2. Dog 97. 9 - 95. 3 94. 8 87. 6 84. 6 66. 6 3. Pig 97. 4 97. 6 - 95. 0 87. 4 87. 2 84. 9 71. 3 4. Ccattle 97. 9 98. 0 97. 9 - 87. 7 87. 5 84. 8 66. 9 5. Mouse 93. 6 93. 1 92. 7 93. 5 - 93. 6 83. 8 68. 0 6. Rat 92. 9 92. 3 92. 1 92. 6 96. 4 - 82. 9 73. 0 7. Chicken 67. 7 67. 8 67. 9 67. 0 66. 4 - 74. 2 8. Zebrafish 51. 5 51. 8 51. 6 51. 4 51. 8 51. 6 - 1. Human
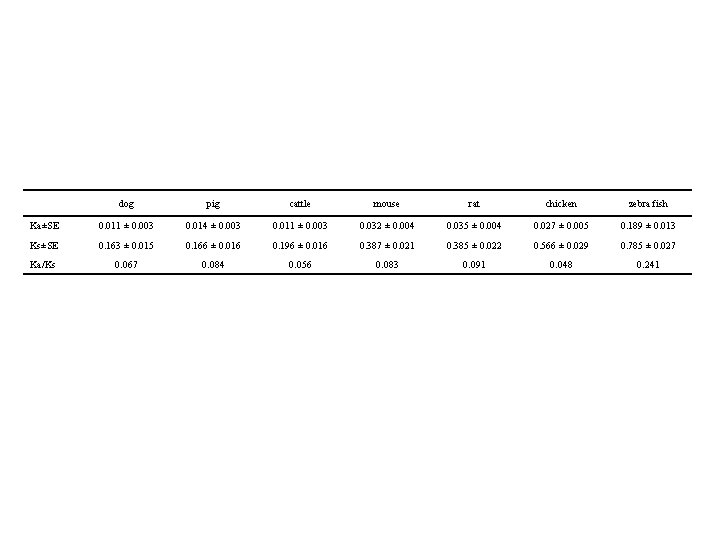
dog pig cattle mouse rat chicken zebra fish Ka±SE 0. 011 ± 0. 003 0. 014 ± 0. 003 0. 011 ± 0. 003 0. 032 ± 0. 004 0. 035 ± 0. 004 0. 027 ± 0. 005 0. 189 ± 0. 013 Ks±SE 0. 163 ± 0. 015 0. 166 ± 0. 016 0. 196 ± 0. 016 0. 387 ± 0. 021 0. 385 ± 0. 022 0. 566 ± 0. 029 0. 785 ± 0. 027 Ka/Ks 0. 067 0. 084 0. 056 0. 083 0. 091 0. 048 0. 241

- Slides: 31