VIROLOGY viruses and nonchromosomal genetic elements VIRAL GENETICS

- Slides: 58

VIROLOGY (viruses and non-chromosomal genetic elements) VIRAL GENETICS

VIRAL GENETICS Mutation types : Biochemical characterization phenotypic expression MUTATION FREQUENCIES OF VIRUSES Interaction between viruses and cells phenotypic mixing Reasortiments Helper viruses Interference restriction-modification CRISP/Cas system The lytic and lysogenic development cycle, immunity Transduction

TYPES OF MUTATION: single nucleotide replacement : transition or transversion misssense, nonsense or silent insertion /deletion of nucleotides recombination genomic mutations: translocations inversions deletions duplications

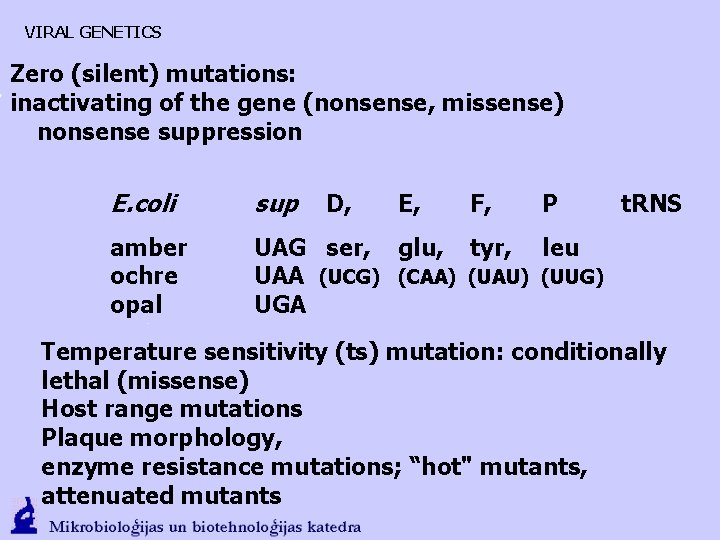
VIRAL GENETICS Zero (silent) mutations: inactivating of the gene (nonsense, missense) nonsense suppression E. coli sup amber ochre opal UAG ser, glu, tyr, leu UAA (UCG) (CAA) (UAU) (UUG) UGA D, E, F, P t. RNS Temperature sensitivity (ts) mutation: conditionally lethal (missense) Host range mutations Plaque morphology, enzyme resistance mutations; “hot" mutants, attenuated mutants

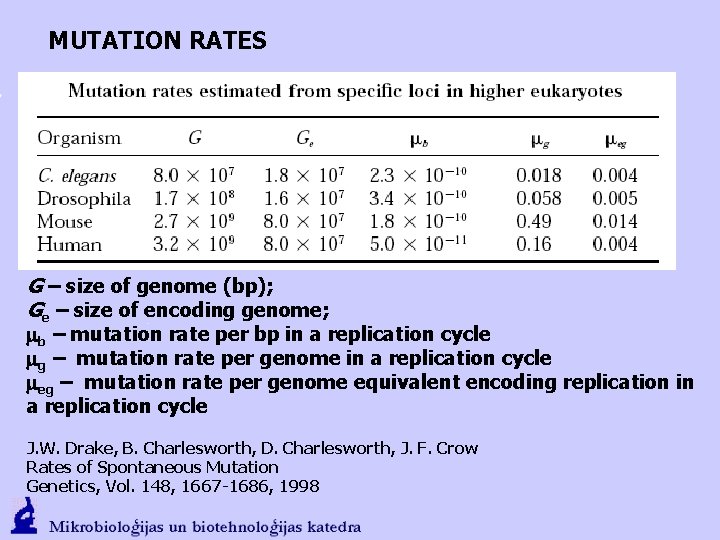
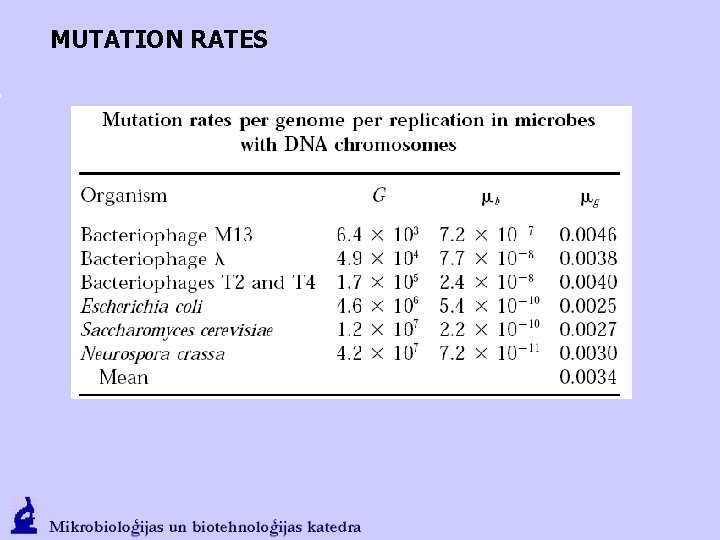
MUTATION RATES G – size of genome (bp); Ge – size of encoding genome; mb – mutation rate per bp in a replication cycle mg – mutation rate per genome in a replication cycle meg – mutation rate per genome equivalent encoding replication in a replication cycle J. W. Drake, B. Charlesworth, D. Charlesworth, J. F. Crow Rates of Spontaneous Mutation Genetics, Vol. 148, 1667 -1686, 1998

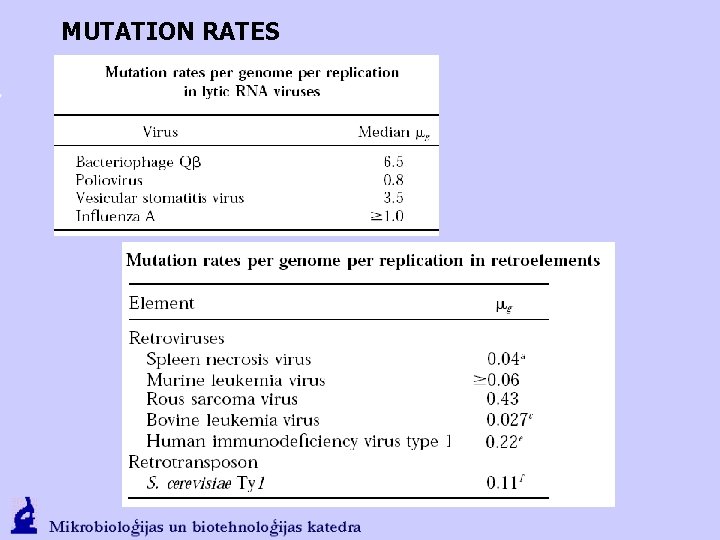
MUTATION RATES

MUTATION RATES

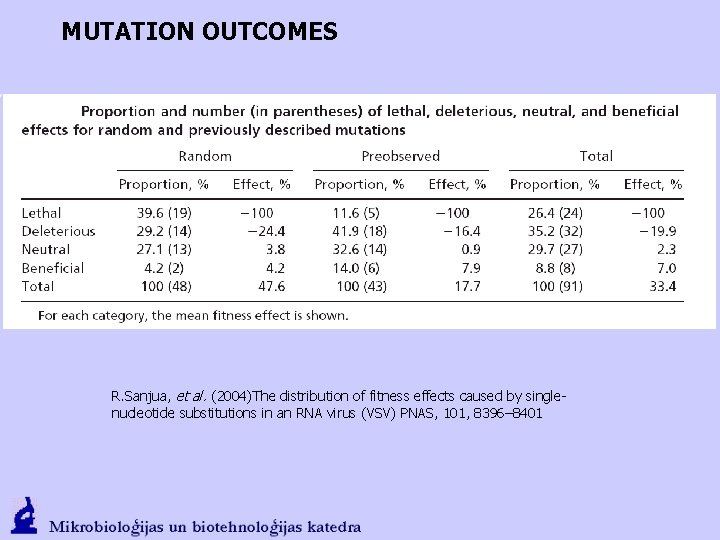
MUTATION OUTCOMES R. Sanjua, et al. (2004)The distribution of fitness effects caused by singlenucleotide substitutions in an RNA virus (VSV) PNAS, 101, 8396– 8401

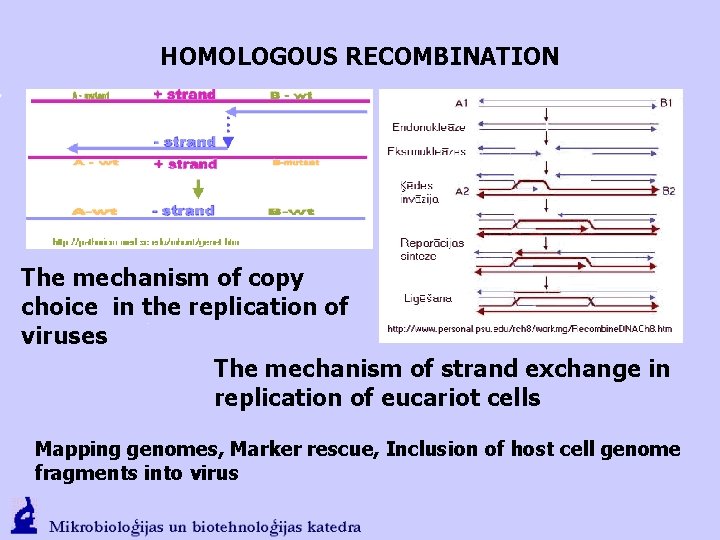
HOMOLOGOUS RECOMBINATION The mechanism of copy choice in the replication of viruses The mechanism of strand exchange in replication of eucariot cells Mapping genomes, Marker rescue, Inclusion of host cell genome fragments into virus

REASSORTMENT of viruses with segmented genome Opportunities for the development of vaccines using the reassortment of influenza virus genome

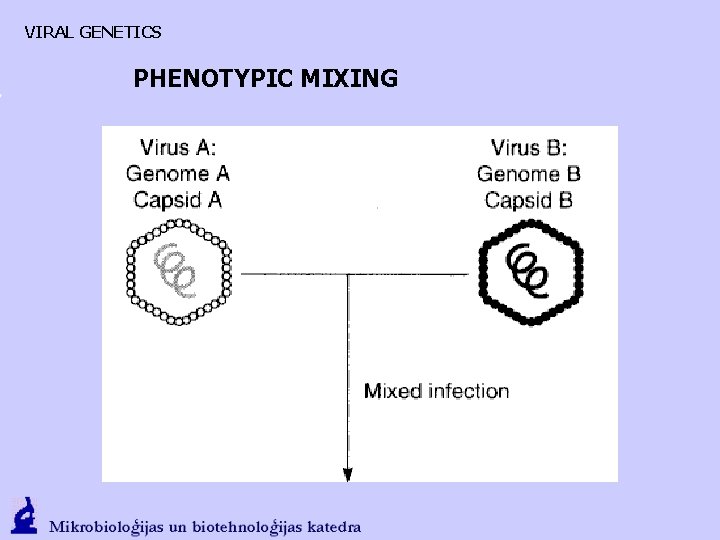
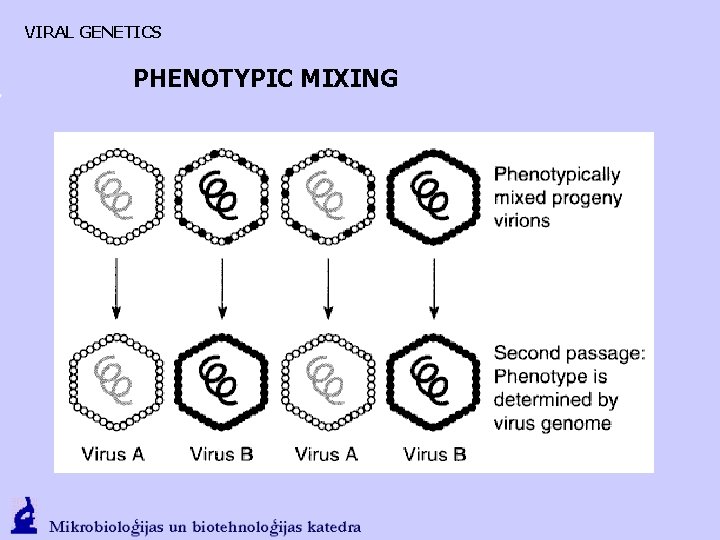
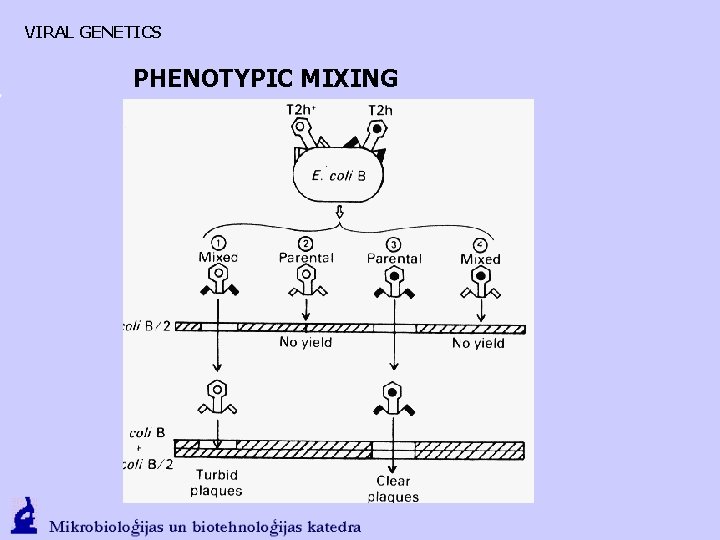
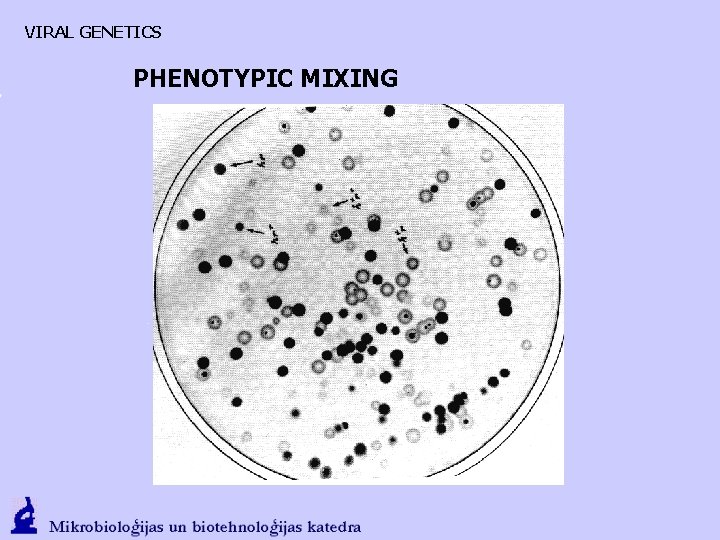
VIRAL GENETICS PHENOTYPIC MIXING

VIRAL GENETICS PHENOTYPIC MIXING

VIRAL GENETICS PHENOTYPIC MIXING

VIRAL GENETICS PHENOTYPIC MIXING

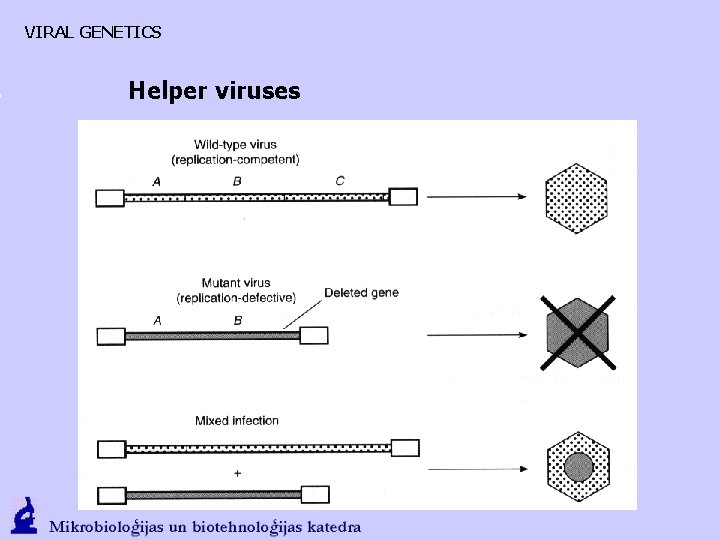
VIRAL GENETICS Helper viruses

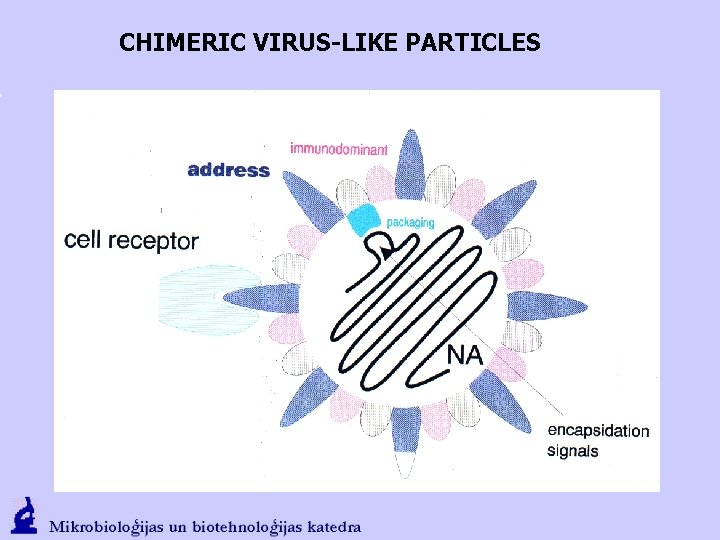
CHIMERIC VIRUS-LIKE PARTICLES

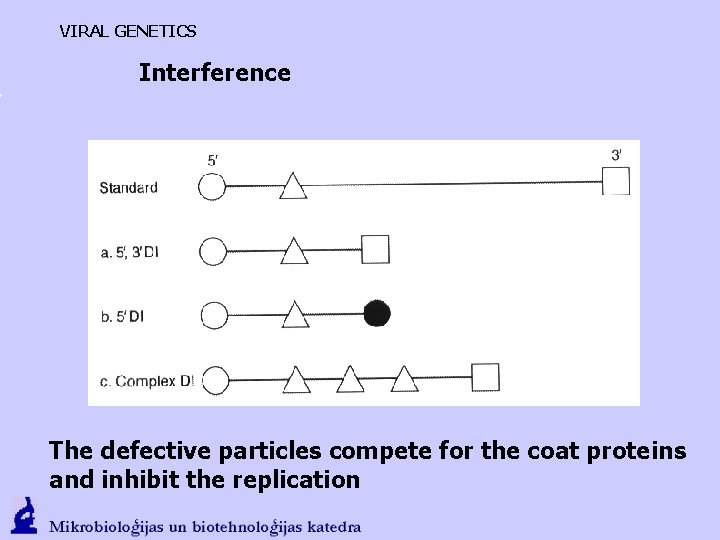
VIRAL GENETICS Interference The defective particles compete for the coat proteins and inhibit the replication

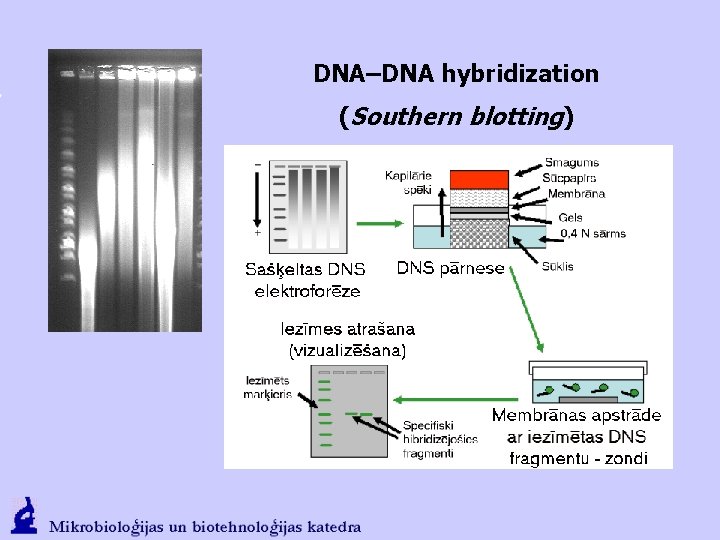
DNA–DNA hybridization (Southern blotting)

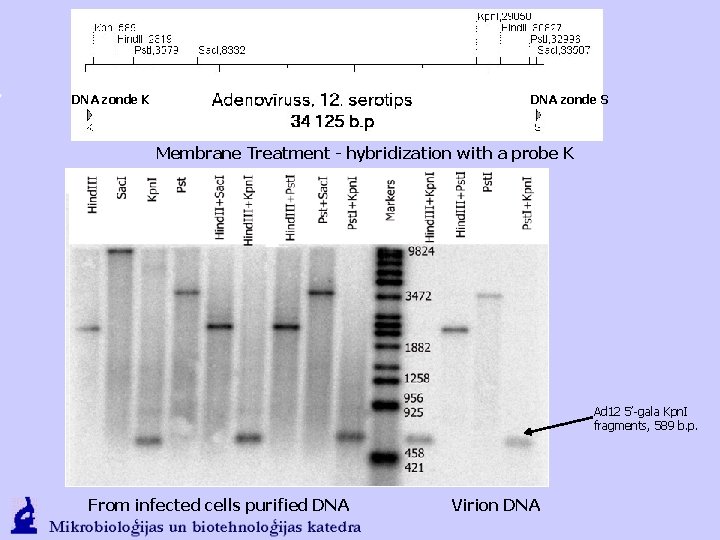
DNA zonde K DNA zonde S Membrane Treatment - hybridization with a probe K Ad 12 5’-gala Kpn. I fragments, 589 b. p. From infected cells purified DNA Virion DNA

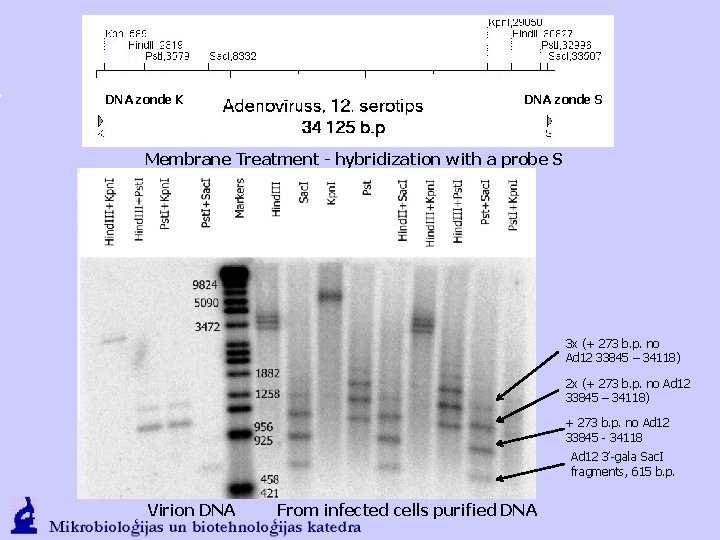
DNA zonde K DNA zonde S Membrane Treatment - hybridization with a probe S 3 x (+ 273 b. p. no Ad 12 33845 – 34118) 2 x (+ 273 b. p. no Ad 12 33845 – 34118) + 273 b. p. no Ad 12 33845 - 34118 Ad 12 3’-gala Sac. I fragments, 615 b. p. Virion DNA From infected cells purified DNA

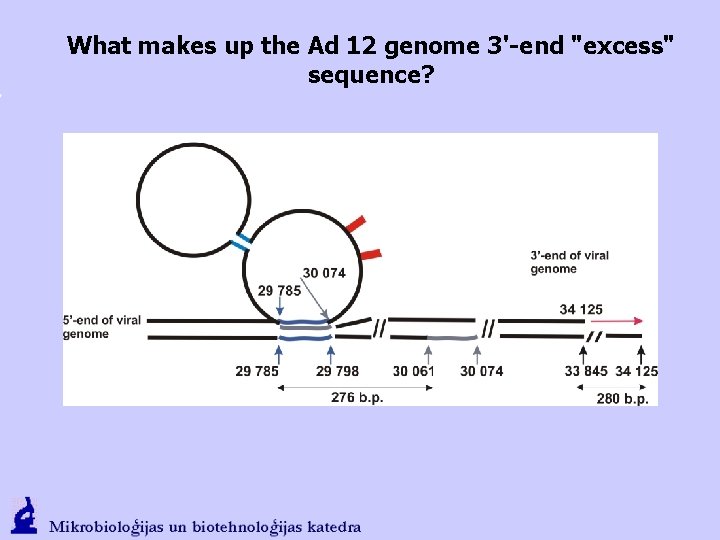
What makes up the Ad 12 genome 3'-end "excess" sequence?

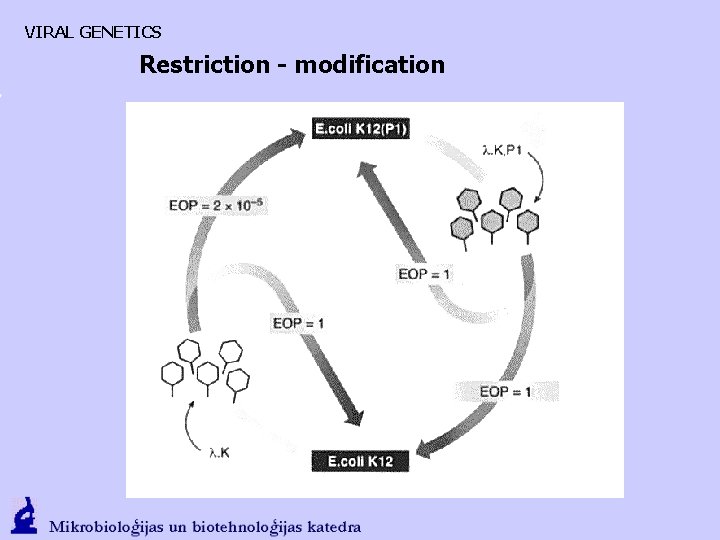
VIRAL GENETICS Restriction - modification

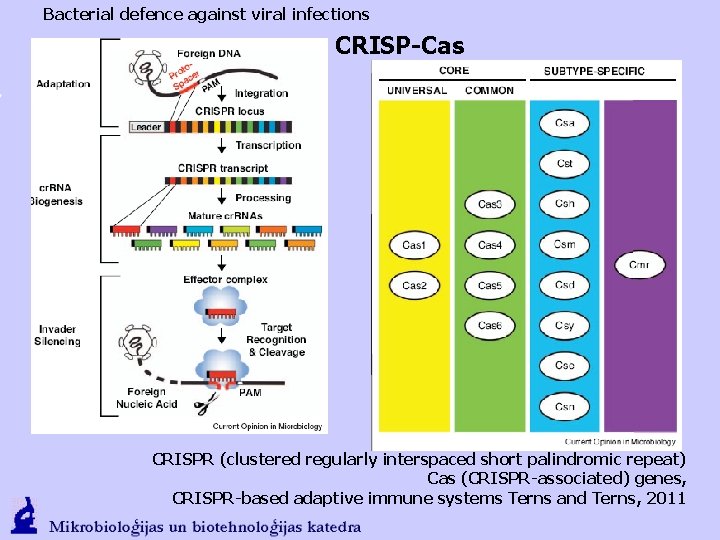
Bacterial defence against viral infections CRISP-Cas CRISPR (clustered regularly interspaced short palindromic repeat) Cas (CRISPR-associated) genes, CRISPR-based adaptive immune systems Terns and Terns, 2011

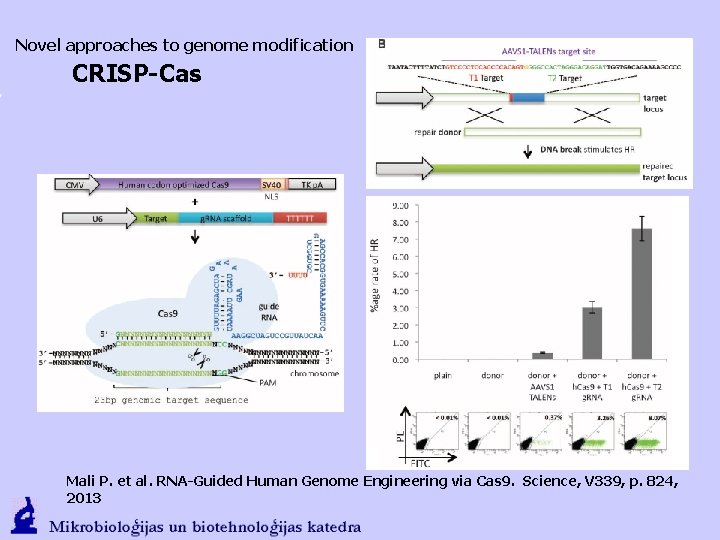
Novel approaches to genome modification CRISP-Cas Mali P. et al. RNA-Guided Human Genome Engineering via Cas 9. Science, V 339, p. 824, 2013

VIRAL GENETICS Transfection Protein unprotected viral delivery of genetic material in the cell (electroporation, liposomes, hydroxyapatite) Transduction Gene transfer with the help of virus Specialized (l phage, gal, bio operons) Non-specific (P 1, P 22 phage, 40 -50 kbp. genomic fragments)

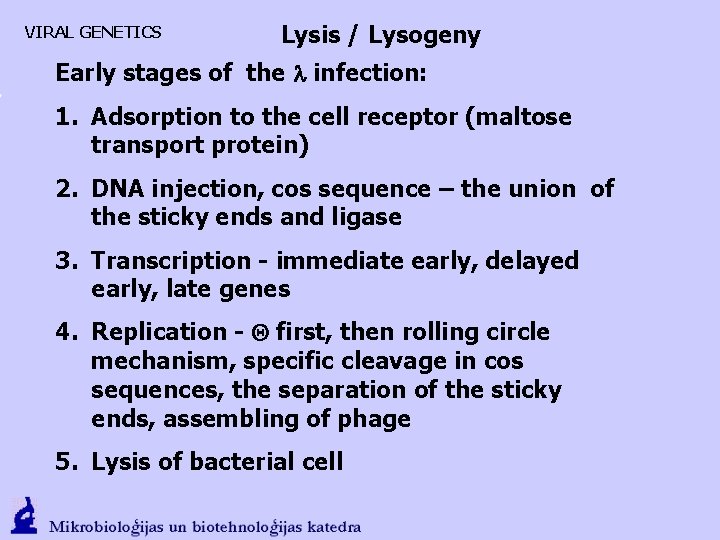
VIRAL GENETICS Lysis / Lysogeny Strategy Choice of the l–phage replication

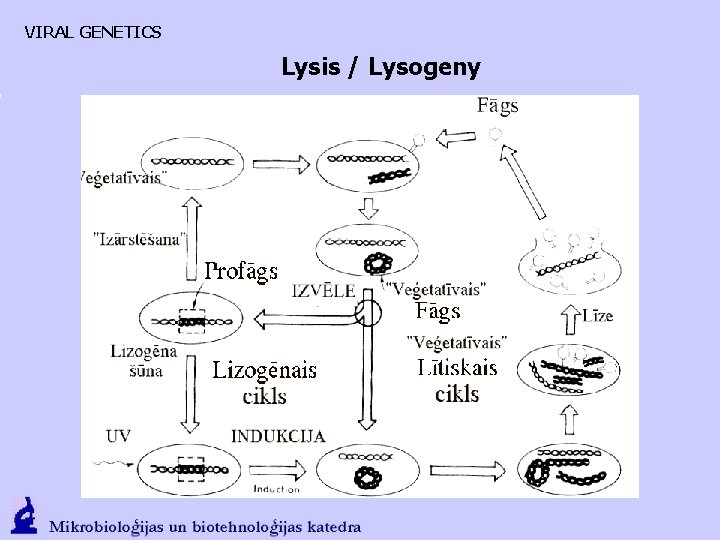
VIRAL GENETICS Lysis / Lysogeny

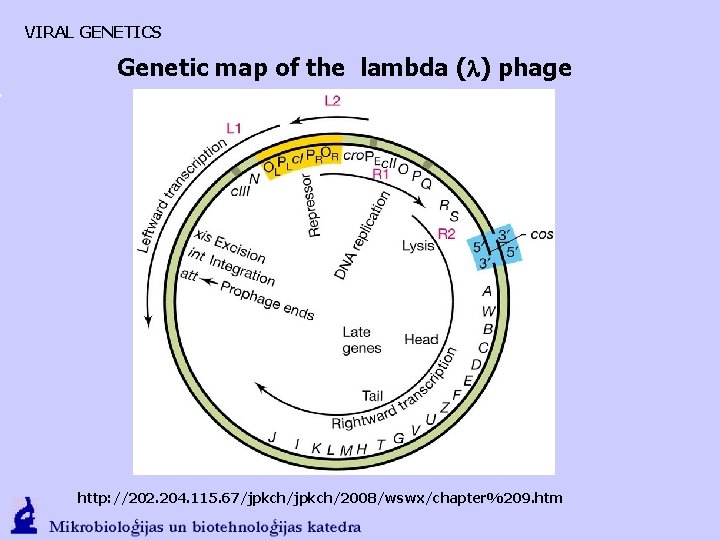
VIRAL GENETICS Genetic map of the lambda (l) phage http: //202. 204. 115. 67/jpkch/2008/wswx/chapter%209. htm

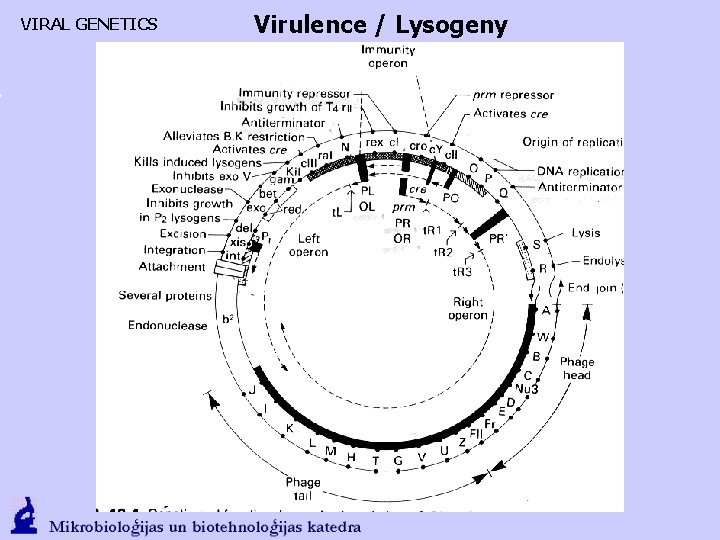
VIRAL GENETICS Virulence / Lysogeny

VIRAL GENETICS Lysis / Lysogeny Early stages of the l infection: 1. Adsorption to the cell receptor (maltose transport protein) 2. DNA injection, cos sequence – the union of the sticky ends and ligase 3. Transcription - immediate early, delayed early, late genes 4. Replication - first, then rolling circle mechanism, specific cleavage in cos sequences, the separation of the sticky ends, assembling of phage 5. Lysis of bacterial cell

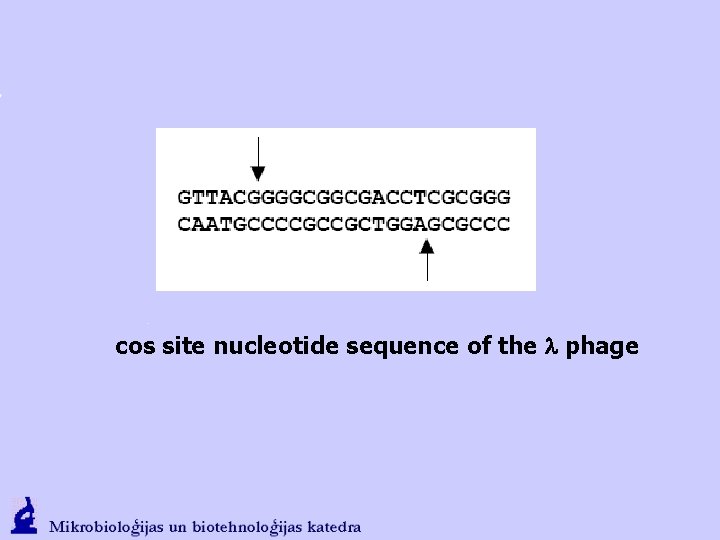
cos site nucleotide sequence of the l phage

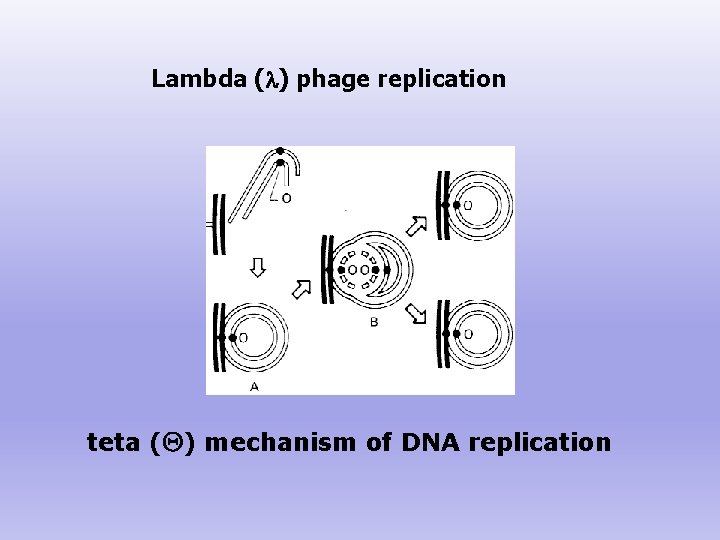
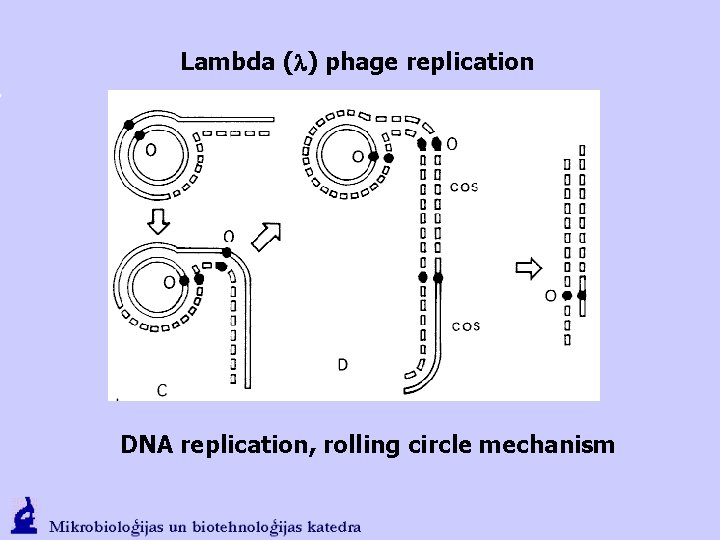
Lambda (l) phage replication teta ( ) mechanism of DNA replication

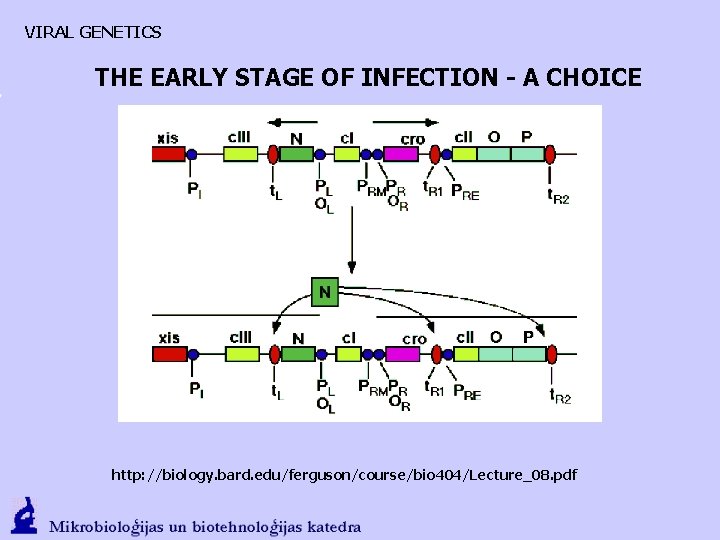
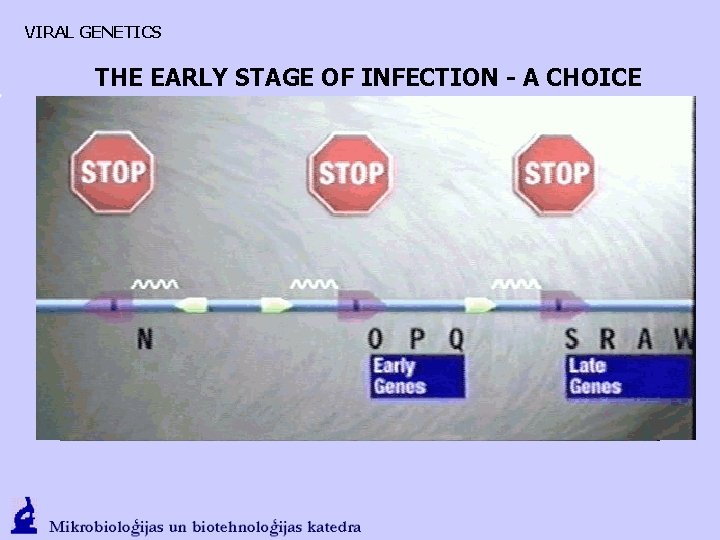
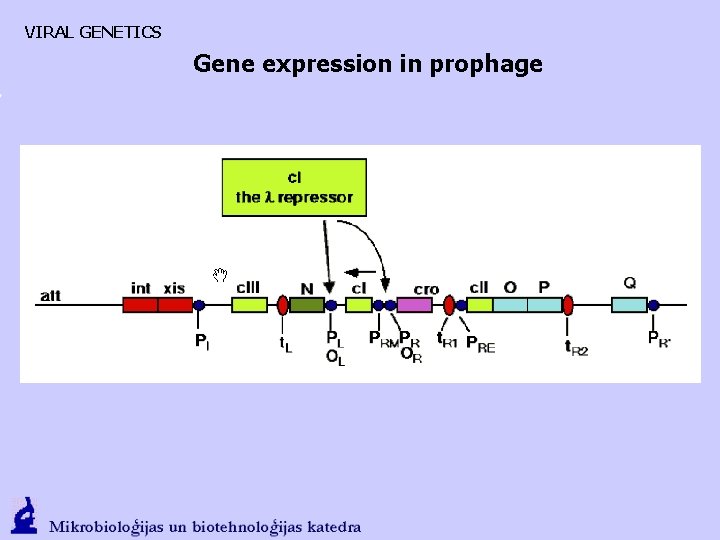
VIRAL GENETICS THE EARLY STAGE OF INFECTION - A CHOICE 1. Weak transcription from PL and PR. Antitermination protein N that interacts with RNA polymerase and promotes transcription in both directions is formed. Cro regulatory protein that promotes transkription of PR is formed. 2. N promotes CIII (CII stabilizer) {PL}; as well as CII (CI stimulator) O, P, (DNA synthesis, mechanism), Q gene transcription {PR}

VIRAL GENETICS THE EARLY STAGE OF INFECTION - A CHOICE http: //biology. bard. edu/ferguson/course/bio 404/Lecture_08. pdf

VIRAL GENETICS THE EARLY STAGE OF INFECTION - A CHOICE

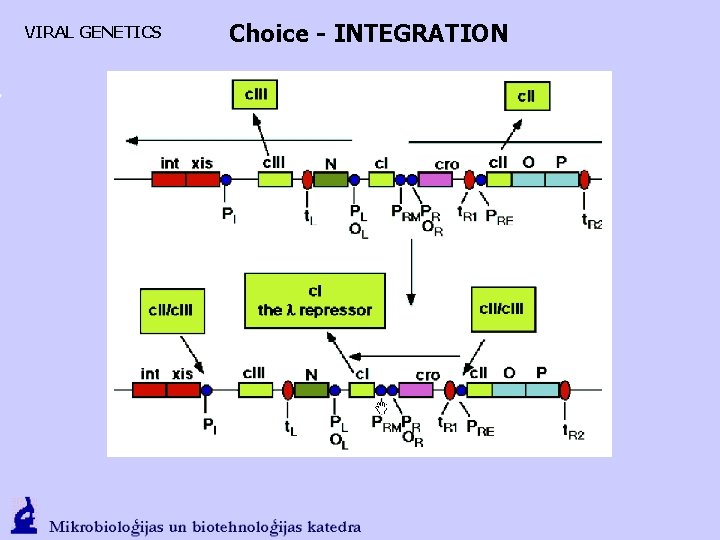
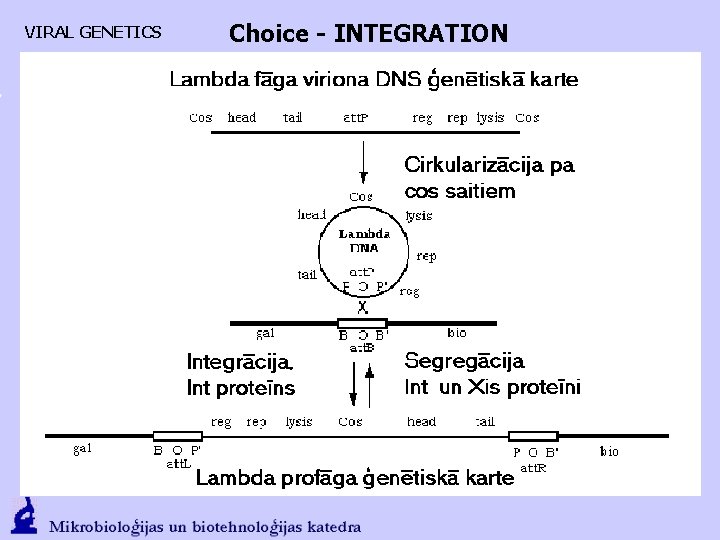
Vīrusu ģenētika Choice - INTEGRATION LYSOGENY. CII activates the PRE (CI synthesis starts) and PI (integrase). Formed CI, which extorts Cro from PL and PR, activates PRM Int promotes att. P and att. B interaction and a fusion of DNA of phage with the DNA of bacteria.

VIRAL GENETICS Choice - INTEGRATION

VIRAL GENETICS Choice - INTEGRATION

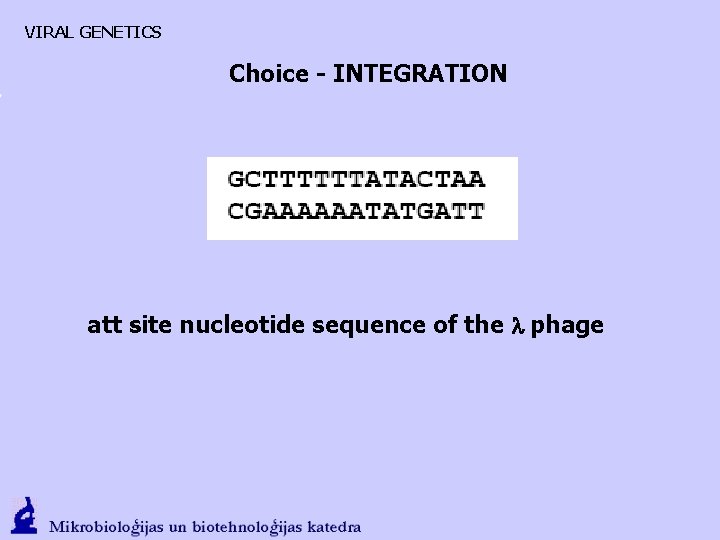
VIRAL GENETICS Choice - INTEGRATION att site nucleotide sequence of the l phage

VIRAL GENETICS Choice - INTEGRATION

VIRAL GENETICS Choice - INTEGRATION

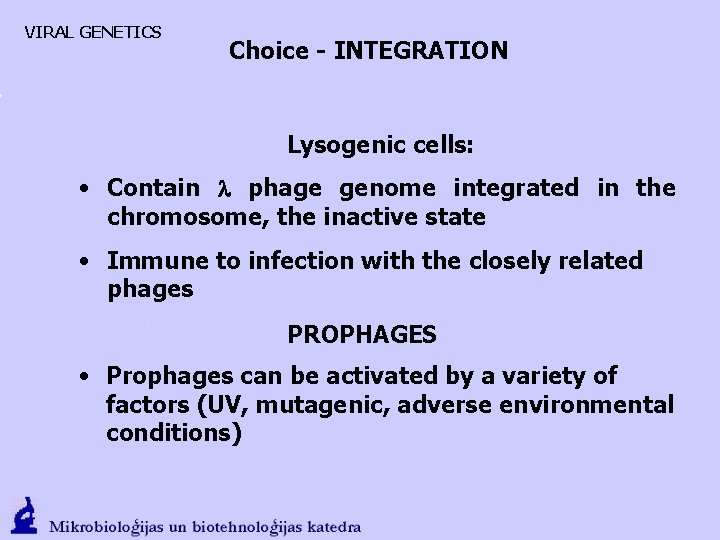
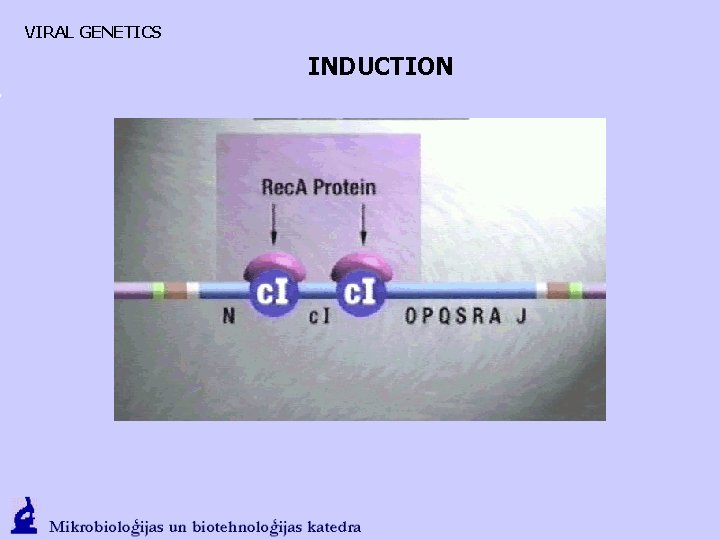
VIRAL GENETICS Choice - INTEGRATION Lysogenic cells: • Contain l phage genome integrated in the chromosome, the inactive state • Immune to infection with the closely related phages PROPHAGES • Prophages can be activated by a variety of factors (UV, mutagenic, adverse environmental conditions)

VIRAL GENETICS Gene expression in prophage

VIRAL GENETICS INDUCTION

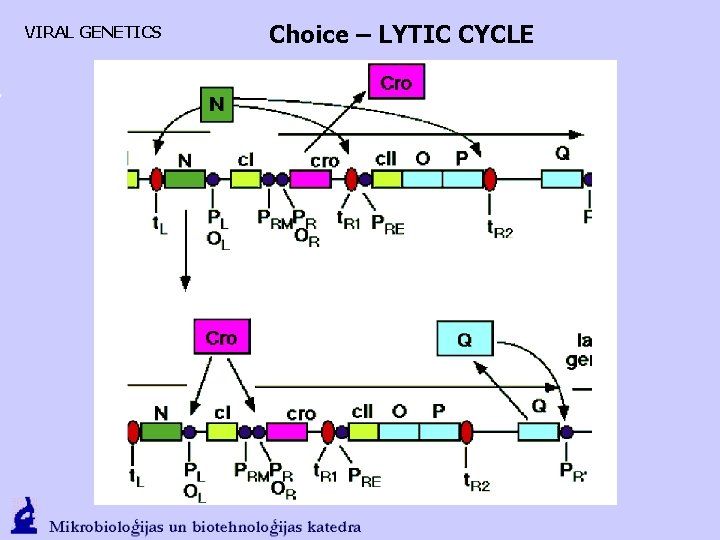
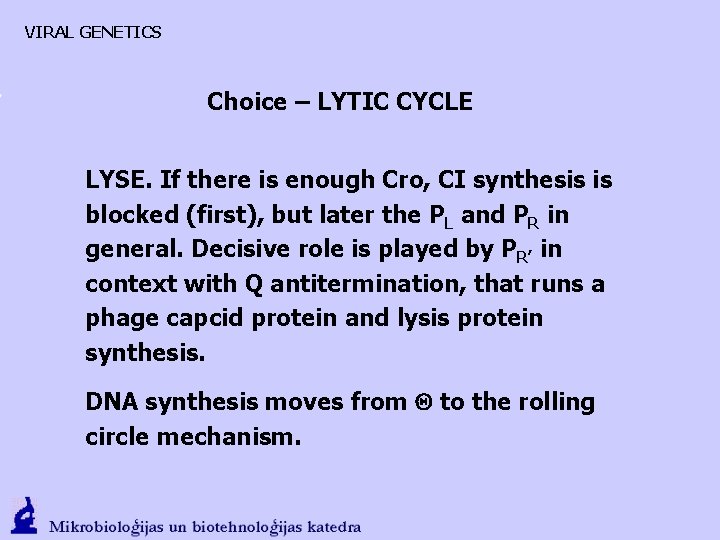
VIRAL GENETICS Choice – LYTIC CYCLE

Lambda (l) phage replication DNA replication, rolling circle mechanism

VIRAL GENETICS Choice – LYTIC CYCLE LYSE. If there is enough Cro, CI synthesis is blocked (first), but later the PL and PR in general. Decisive role is played by PR’ in context with Q antitermination, that runs a phage capcid protein and lysis protein synthesis. DNA synthesis moves from to the rolling circle mechanism.

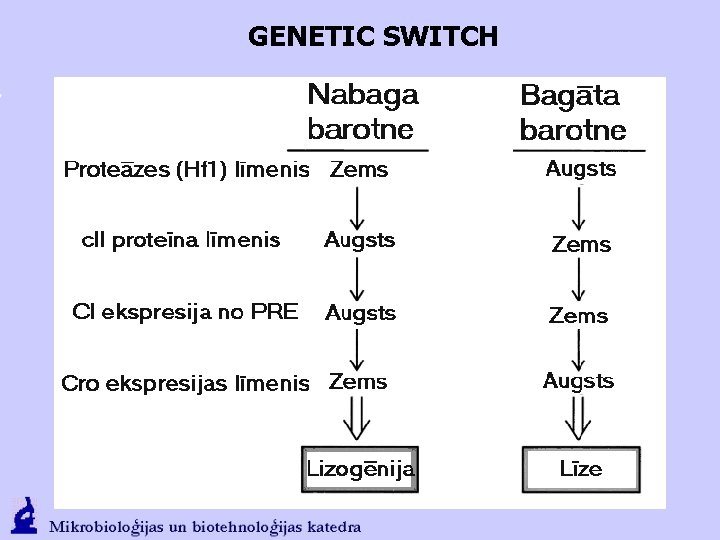
GENETIC SWITCH

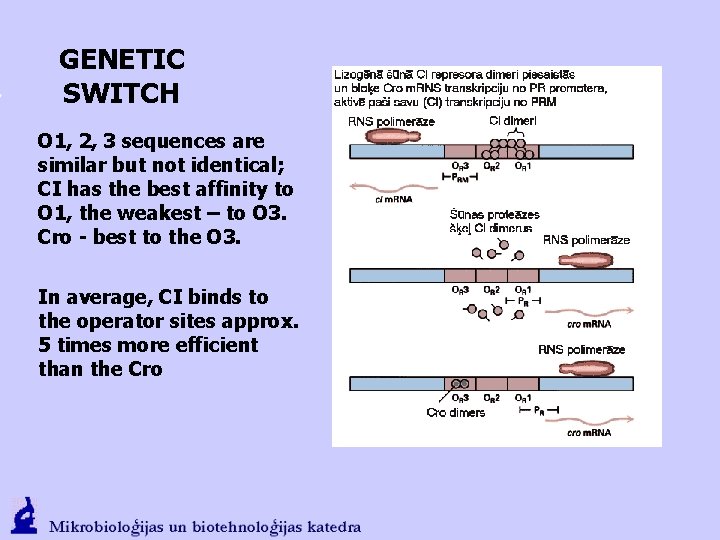
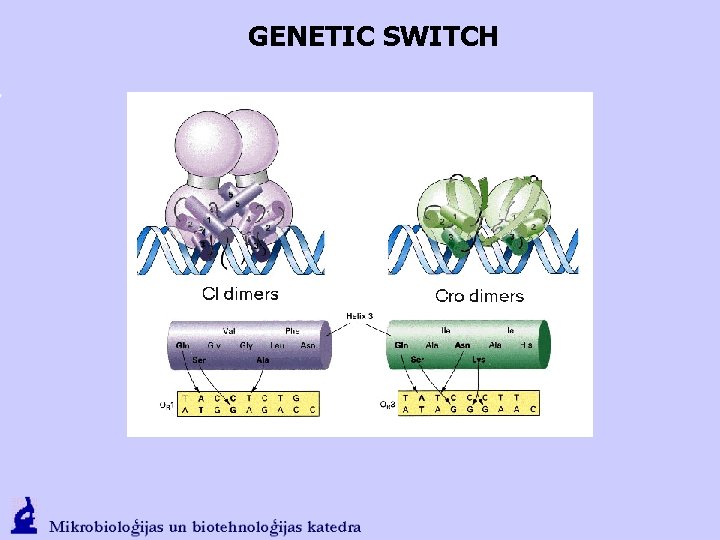
GENETIC SWITCH O 1, 2, 3 sequences are similar but not identical; CI has the best affinity to O 1, the weakest – to O 3. Cro - best to the O 3. In average, CI binds to the operator sites approx. 5 times more efficient than the Cro

GENETIC SWITCH

OTHER E. coli LYSOGENE PHAGES • l phage-like – phages 21 f 80, 82, 424, 434, crossimmunity; • P 1, the largest lysogene phage, 97 kbp. DNA rarely integrates - more present in plasmid form of Cre protein and lox. P recombination site, 40% of the DNA filling required for aggregation, non-specific transduction; • Mu, 42 kbp. DNA, at the ends of phage genome – bacteria sequence, effective transposon, mutation induction; • P 2, 33, 2 kbp. DNA, approx. 10 integration sites in the genome of bacteria, lysis is rare. P 2 encoded capsid proteins can be used for P 4 (11 kpb. DNA) incapsidation, which in P 2 free cells are in multicopy plasmid form

VIRAL GENETICS TRANSDUCTION Gene transfer with the help of LYSOGENE virus Specialized (l phage, gal, bio operons) Non-specific (P 2 phage, 40 -50 KBP. genomic fragments)

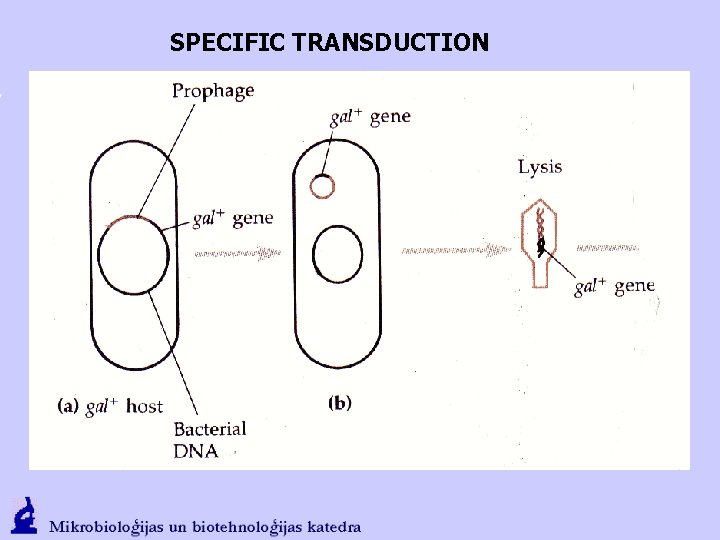
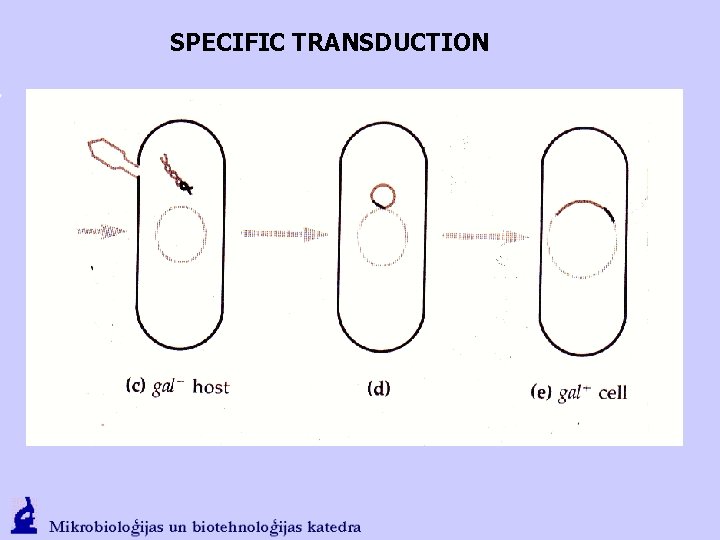
SPECIFIC TRANSDUCTION

SPECIFIC TRANSDUCTION

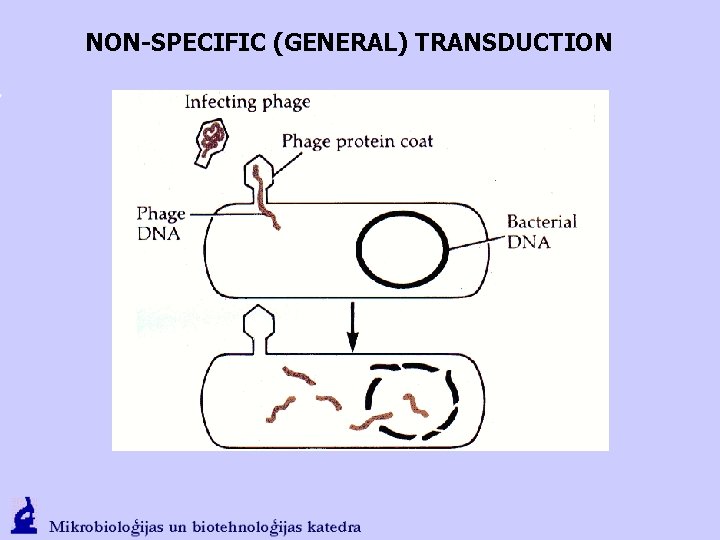
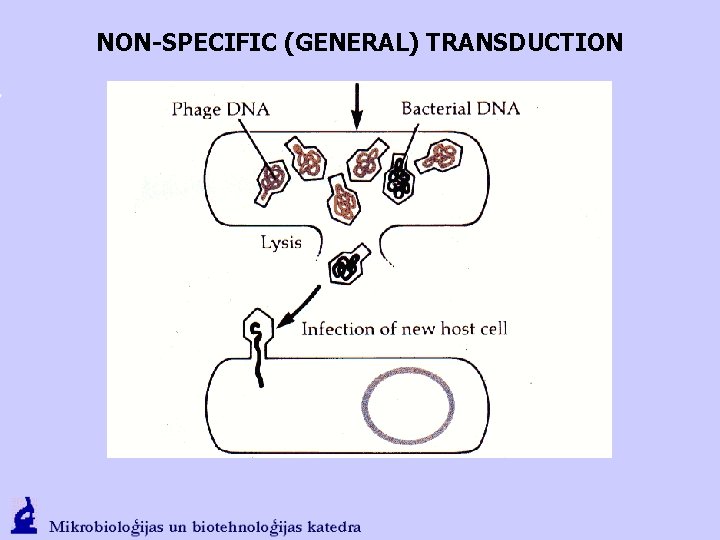
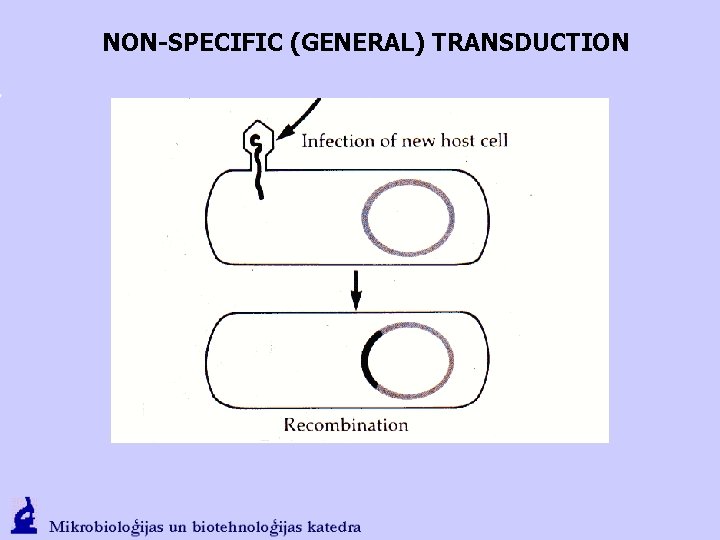
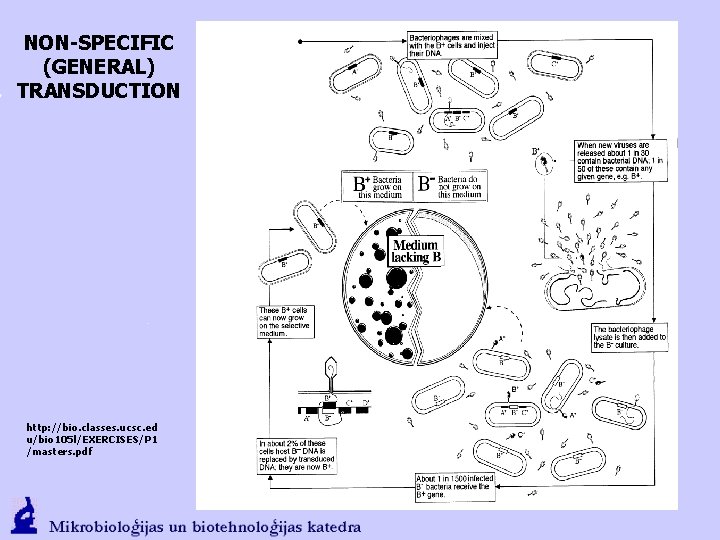
NON-SPECIFIC (GENERAL) TRANSDUCTION

NON-SPECIFIC (GENERAL) TRANSDUCTION

NON-SPECIFIC (GENERAL) TRANSDUCTION

NON-SPECIFIC (GENERAL) TRANSDUCTION http: //bio. classes. ucsc. ed u/bio 105 l/EXERCISES/P 1 /masters. pdf
 Https://m..com/watch?v=nqecciuc7jy
Https://m..com/watch?v=nqecciuc7jy Unlike lytic viruses, lysogenic viruses do not
Unlike lytic viruses, lysogenic viruses do not Introduction to medical virology
Introduction to medical virology Duhare
Duhare Virology
Virology Fields virology
Fields virology Fields virology
Fields virology Chapter 12 section 1 dna the genetic material
Chapter 12 section 1 dna the genetic material Founder effect
Founder effect Genetic programming vs genetic algorithm
Genetic programming vs genetic algorithm Genetic programming vs genetic algorithm
Genetic programming vs genetic algorithm Genetic drift vs gene flow
Genetic drift vs gene flow What is the difference between genetic drift and gene flow
What is the difference between genetic drift and gene flow Section 24-1 viral structure and replication
Section 24-1 viral structure and replication Section 19-3 diseases caused by bacteria and viruses
Section 19-3 diseases caused by bacteria and viruses Egrette chapter 21
Egrette chapter 21 Nonliving particle that replicates inside a living cell
Nonliving particle that replicates inside a living cell What are viruses lesson 3 answer key
What are viruses lesson 3 answer key Chapter 20 viruses and prokaryotes
Chapter 20 viruses and prokaryotes Study guide chapter 18 section 1 bacteria
Study guide chapter 18 section 1 bacteria Section 1 studying viruses and prokaryotes
Section 1 studying viruses and prokaryotes Viral induced wheeze vs asthma
Viral induced wheeze vs asthma Vrus
Vrus Pantropizm
Pantropizm Method of cultivation of virus
Method of cultivation of virus Viral inoculation in embryonated egg
Viral inoculation in embryonated egg Egg inoculation diagram
Egg inoculation diagram Spasmodic croup vs viral croup
Spasmodic croup vs viral croup Variola varicela
Variola varicela Exocitose
Exocitose When blood sample hemolyzed
When blood sample hemolyzed Meningitis
Meningitis Causes of viral hemorrhagic fever
Causes of viral hemorrhagic fever Ciclo viral
Ciclo viral Capsid capsomere
Capsid capsomere Viral
Viral Viral recombination
Viral recombination Vaccins à vecteur viral
Vaccins à vecteur viral Viral receptors
Viral receptors Streptococcus
Streptococcus Trivalent vaccin herpes
Trivalent vaccin herpes Viral dna
Viral dna Rotarix live attenuated
Rotarix live attenuated Viral entry
Viral entry The dynamics of viral marketing
The dynamics of viral marketing Hepatitis viral
Hepatitis viral Csf analysis interpretation
Csf analysis interpretation Viral arthritis
Viral arthritis An acute highly contagious viral disease
An acute highly contagious viral disease Viral life cycle
Viral life cycle Mobilivirus
Mobilivirus Viral shedding
Viral shedding Eline's viral
Eline's viral Viral communications
Viral communications Viral integration
Viral integration Vacinas subcutânea
Vacinas subcutânea Why are viruses considered nonliving?
Why are viruses considered nonliving? General characteristics of viruses
General characteristics of viruses General characteristics of viruses
General characteristics of viruses