Triosephosphate isomerase EC 5 3 1 1 TIM



































![BLASTP 2. 2. 1 [Apr-13 -2001] BLASTの 出力例(1) Reference: Altschul, Stephen F. , Thomas BLASTP 2. 2. 1 [Apr-13 -2001] BLASTの 出力例(1) Reference: Altschul, Stephen F. , Thomas](https://slidetodoc.com/presentation_image_h/14e21021e916e7c1c34a6bdf78c8a5c7/image-44.jpg)
![1 b 15 A [c. 2. 1. 2] ALCOHOL DEHYDROGENASE 1 pmi- [b. 82. 1 b 15 A [c. 2. 1. 2] ALCOHOL DEHYDROGENASE 1 pmi- [b. 82.](https://slidetodoc.com/presentation_image_h/14e21021e916e7c1c34a6bdf78c8a5c7/image-45.jpg)




![BLASTP 2. 2. 1 [Apr-13 -2001] Reference: Altschul, Stephen F. , Thomas L. Madden, BLASTP 2. 2. 1 [Apr-13 -2001] Reference: Altschul, Stephen F. , Thomas L. Madden,](https://slidetodoc.com/presentation_image_h/14e21021e916e7c1c34a6bdf78c8a5c7/image-50.jpg)

![blastp(アミノ酸対アミノ酸)によるタンパク質の機能予測 クエリ:T. thermophiusのタンパク質, データベース:大腸菌の全タンパク質 BLASTP 2. 2. 3 [May-13 -2002] Query= X 07 AAS blastp(アミノ酸対アミノ酸)によるタンパク質の機能予測 クエリ:T. thermophiusのタンパク質, データベース:大腸菌の全タンパク質 BLASTP 2. 2. 3 [May-13 -2002] Query= X 07 AAS](https://slidetodoc.com/presentation_image_h/14e21021e916e7c1c34a6bdf78c8a5c7/image-56.jpg)


- Slides: 58







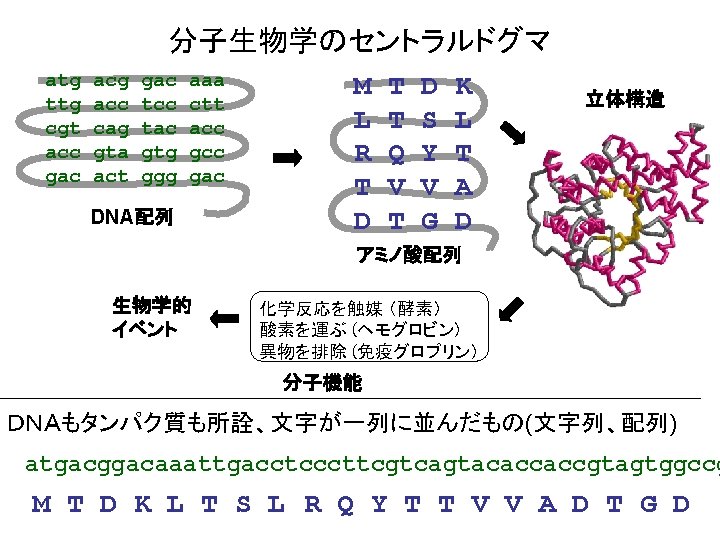
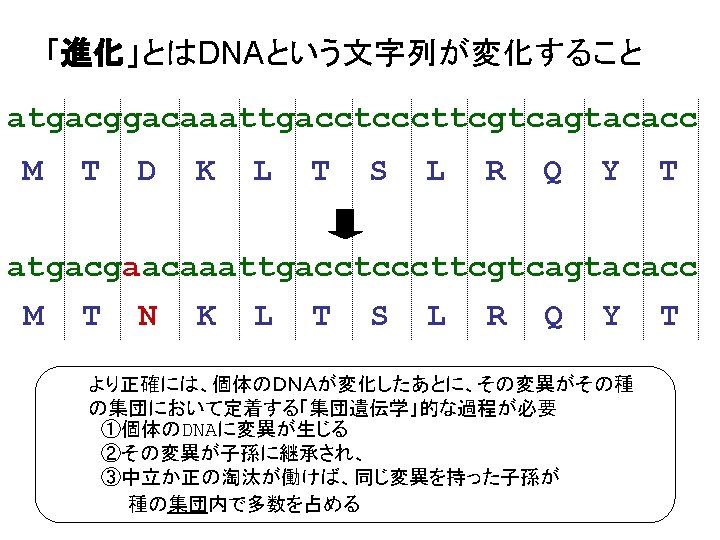
違う生物の同じ機能のタンパク質のアミノ酸配列 トリオースリン酸異性化酵素( Triosephosphate isomerase (EC 5. 3. 1. 1) (TIM, TPIS)) >TPIS_HUMAN ヒト "Triosephosphate isomerase (EC 5. 3. APSRKFFVGGNWKMNGRKQSLGELIGTLNAAKVPADTEVVCAPPT AYIDFARQKLDPKIAVAAQNCYKVTNGAFTGEISPGMIKDCGATW VVLGHSERRHVFGESDELIGQKVAHALAEGLGVIACIGEKLDERE AGITEKVVFEQTKVIADNVKDWSKVVLAYEPVWAIGTGKTATPQQ AQEVHEKLRGWLKSNVSDAVAQSTRIIYGGSVTGATCKELASQPD VDGFLVGGASLKPEFVDIINAKQ >TPIS_RABIT ウサギ "Triosephosphate isomerase (EC 5 APSRKFFVGGNWKMNGRKKNLGELITTLNAAKVPADTEVVCAPPT AYIDFARQKLDPKIAVAAQNCYKVTNGAFTGEISPGMIKDCGATW VVLGHSERRHVFGESDELIGQKVAHALSEGLGVIACIGEKLDERE AGITEKVVFEQTKVIADNVKDWSKVVLAYEPVWAIGTGKTATPQQ AQEVHEKLRGWLKSNVSDAVAQSTRIIYGGSVTGATCKELASQPD VDGFLVGGASLKPEFVDIINAKQ

違う生物の同じ機能のタンパク質のアミノ酸配列 トリオースリン酸異性化酵素( Triosephosphate isomerase (EC 5. 3. 1. 1) (TIM, TPIS)) >TPIS_HUMAN ヒト "Triosephosphate isomerase (EC 5. 3. APSRKFFVGGNWKMNGRKQSLGELIGTLNAAKVPADTEVVCAPPT AYIDFARQKLDPKIAVAAQNCYKVTNGAFTGEISPGMIKDCGATW VVLGHSERRHVFGESDELIGQKVAHALAEGLGVIACIGEKLDERE AGITEKVVFEQTKVIADNVKDWSKVVLAYEPVWAIGTGKTATPQQ AQEVHEKLRGWLKSNVSDAVAQSTRIIYGGSVTGATCKELASQPD VDGFLVGGASLKPEFVDIINAKQ >TPIS_YEAST 酵母 "Triosephosphate isomerase (EC 5. 3 ARTFFVGGNFKLNGSKQSIKEIVERLNTASIPENVEVVICPPATY LDYSVSLVKKPQVTVGAQNAYLKASGAFTGENSVDQIKDVGAKWV ILGHSERRSYFHEDDKFIADKTKFALGQGVGVILCIGETLEEKKA GKTLDVVERQLNAVLEEVKDWTNVVVAYEPVWAIGTGLAATPEDA QDIHASIRKFLASKLGDKAASELRILYGGSANGSNAVTFKDKADV DGFLVGGASLKPEFVDIINSRN

違う生物の同じ機能のタンパク質のアミノ酸配列 トリオースリン酸異性化酵素( Triosephosphate isomerase (EC 5. 3. 1. 1) (TIM, TPIS)) >TPIS_HUMAN ヒト "Triosephosphate isomerase (EC 5. 3. APSRKFFVGGNWKMNGRKQSLGELIGTLNAAKVPADTEVVCAPPT AYIDFARQKLDPKIAVAAQNCYKVTNGAFTGEISPGMIKDCGATW VVLGHSERRHVFGESDELIGQKVAHALAEGLGVIACIGEKLDERE AGITEKVVFEQTKVIADNVKDWSKVVLAYEPVWAIGTGKTATPQQ AQEVHEKLRGWLKSNVSDAVAQSTRIIYGGSVTGATCKELASQPD VDGFLVGGASLKPEFVDIINAKQ >TPIS_ECOLI 大腸菌 "Triosephosphate isomerase (EC 5 MRHPLVMGNWKLNGSRHMVHELVSNLRKELAGVAGCAVAIAPPEM YIDMAKREAEGSHIMLGAQNVDLNLSGAFTGETSAAMLKDIGAQY IIIGHSERRTYHKESDELIAKKFAVLKEQGLTPVLCIGETEAENE AGKTEEVCARQIDAVLKTQGAAAFEGAVIAYEPVWAIGTGKSATP AQAQAVHKFIRDHIAKVDANIAEQVIIQYGGSVNASNAAELFAQP DIDGALVGGASLKADAFAVIVKAAEAAKQA

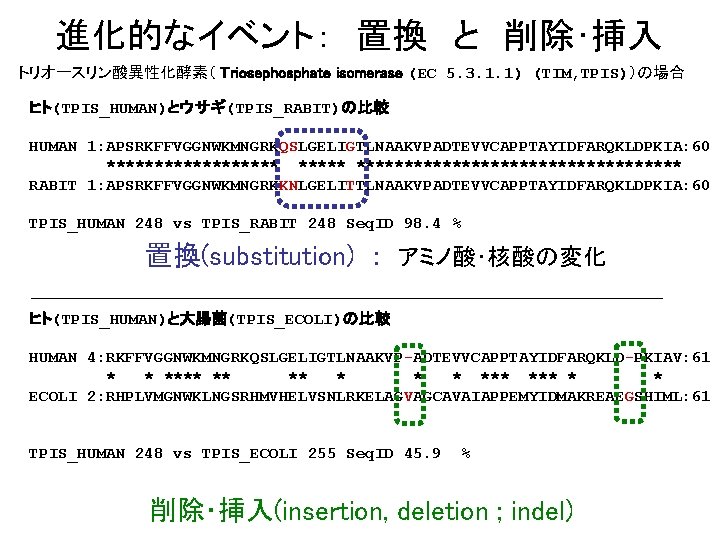
進化的なイベント: 置換 と 削除・挿入 トリオースリン酸異性化酵素( Triosephosphate isomerase (EC 5. 3. 1. 1) (TIM, TPIS))の場合 ヒト(TPIS_HUMAN)とウサギ(TPIS_RABIT)の比較 HUMAN 1: APSRKFFVGGNWKMNGRKQSLGELIGTLNAAKVPADTEVVCAPPTAYIDFARQKLDPKIA: 60 ******************** RABIT 1: APSRKFFVGGNWKMNGRKKNLGELITTLNAAKVPADTEVVCAPPTAYIDFARQKLDPKIA: 60 TPIS_HUMAN 248 vs TPIS_RABIT 248 Seq. ID 98. 4 % 置換(substitution) : アミノ酸・核酸の変化 ヒト(TPIS_HUMAN)と大腸菌(TPIS_ECOLI)の比較 HUMAN 4: RKFFVGGNWKMNGRKQSLGELIGTLNAAKVP-ADTEVVCAPPTAYIDFARQKLD-PKIAV: 61 * * **** ** *** * * ECOLI 2: RHPLVMGNWKLNGSRHMVHELVSNLRKELAGVAGCAVAIAPPEMYIDMAKREAEGSHIML: 61 TPIS_HUMAN 248 vs TPIS_ECOLI 255 Seq. ID 45. 9 % 削除・挿入(insertion, deletion ; indel)

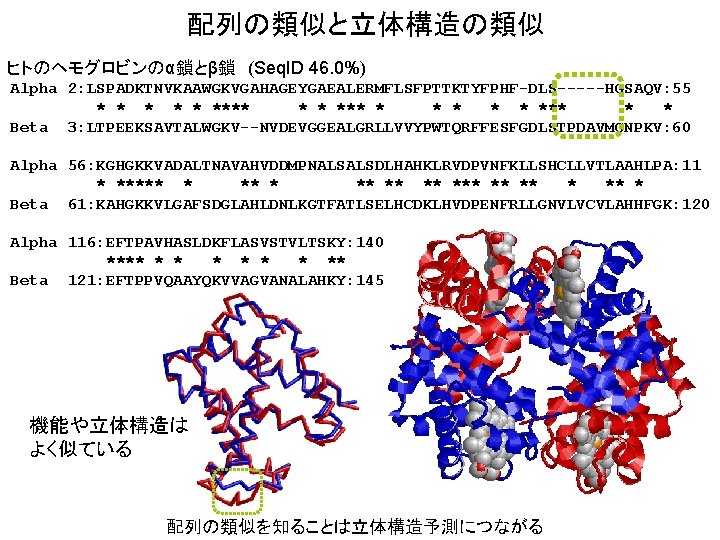
配列の類似と立体構造の類似 ヒトのヘモグロビンのα鎖とβ鎖 (Seq. ID 46. 0%) Alpha 2: LSPADKTNVKAAWGKVGAHAGEYGAEALERMFLSFPTTKTYFPHF-DLS-----HGSAQV: 55 * * **** * * *** * * Beta 3: LTPEEKSAVTALWGKV--NVDEVGGEALGRLLVVYPWTQRFFESFGDLSTPDAVMGNPKV: 60 Alpha 56: KGHGKKVADALTNAVAHVDDMPNALSALSDLHAHKLRVDPVNFKLLSHCLLVTLAAHLPA: 11 * ***** ** ** * Beta 61: KAHGKKVLGAFSDGLAHLDNLKGTFATLSELHCDKLHVDPENFRLLGNVLVCVLAHHFGK: 120 Alpha 116: EFTPAVHASLDKFLASVSTVLTSKY: 140 **** * * * ** Beta 121: EFTPPVQAAYQKVVAGVANALAHKY: 145 機能や立体構造は よく似ている 配列の類似を知ることは立体構造予測につながる






# A R N D C Q E G H I L K M F P S T W Y V B Z X * BLOSUM 62 (blastpのデフォルトで使われている置換スコア行列) A 4 -1 -2 -2 0 -1 -1 0 -2 -1 -1 -2 -1 1 0 -3 -2 0 -2 -1 0 -4 R -1 5 0 -2 -3 1 0 -2 0 -3 -2 2 -1 -3 -2 -1 -1 -3 -2 -3 -1 0 -1 -4 N -2 0 6 1 -3 0 0 0 1 -3 -3 0 -2 -3 -2 1 0 -4 -2 -3 3 0 -1 -4 D -2 -2 1 6 -3 0 2 -1 -1 -3 -4 -1 -3 -3 -1 0 -1 -4 -3 -3 4 1 -1 -4 C 0 -3 -3 -3 9 -3 -4 -3 -3 -1 -1 -3 -1 -2 -3 -1 -1 -2 -2 -1 -3 -3 -2 -4 Q -1 1 0 0 -3 5 2 -2 0 -3 -2 1 0 -3 -1 0 -1 -2 0 3 -1 -4 E -1 0 0 2 -4 2 5 -2 0 -3 -3 1 -2 -3 -1 0 -1 -3 -2 -2 1 4 -1 -4 G 0 -2 0 -1 -3 -2 -2 6 -2 -4 -4 -2 -3 -3 -2 0 -2 -2 -3 -3 -1 -2 -1 -4 H -2 0 1 -1 -3 0 0 -2 8 -3 -3 -1 -2 -2 2 -3 0 0 -1 -4 I -1 -3 -3 -3 -1 -3 -3 -4 -3 4 2 -3 1 0 -3 -2 -1 -3 -1 3 -3 -3 -1 -4 L -1 -2 -3 -4 -3 2 4 -2 2 0 -3 -2 -1 1 -4 -3 -1 -4 K -1 2 0 -1 -3 1 1 -2 -1 -3 -2 5 -1 -3 -1 0 -1 -3 -2 -2 0 1 -1 -4 M -1 -1 -2 -3 -1 0 -2 -3 -2 1 2 -1 5 0 -2 -1 -1 1 -3 -1 -1 -4 F -2 -3 -3 -3 -1 0 0 -3 0 6 -4 -2 -2 1 3 -1 -3 -3 -1 -4 P -1 -2 -2 -1 -3 -1 -1 -2 -2 -3 -3 -1 -2 -4 7 -1 -1 -4 -3 -2 -2 -1 -2 -4 S 1 -1 1 0 -1 0 0 0 -1 -2 -2 0 -1 -2 -1 4 1 -3 -2 -2 0 0 0 -4 T 0 -1 -1 -2 -2 -1 -1 -2 -1 1 5 -2 -2 0 -1 -1 0 -4 W -3 -3 -4 -4 -2 -2 -3 -1 1 -4 -3 -2 11 2 -3 -4 -3 -2 -4 Y -2 -2 -2 -3 -2 -1 -2 -3 2 -1 -1 -2 -1 3 -3 -2 -2 2 7 -1 -3 -2 -1 -4 V 0 -3 -3 -3 -1 -2 -2 -3 -3 3 1 -2 1 -1 -2 -2 0 -3 -1 4 -3 -2 -1 -4 B -2 -1 3 4 -3 0 1 -1 0 -3 -4 0 -3 -3 -2 0 -1 -4 -3 -3 4 1 -1 -4 Z -1 0 0 1 -3 3 4 -2 0 -3 -3 1 -1 -3 -1 0 -1 -3 -2 -2 1 4 -1 -4 X 0 -1 -1 -1 -2 -1 -1 -1 -2 0 0 -2 -1 -1 -1 -4 * -4 -4 -4 -4 -4 -4 1


















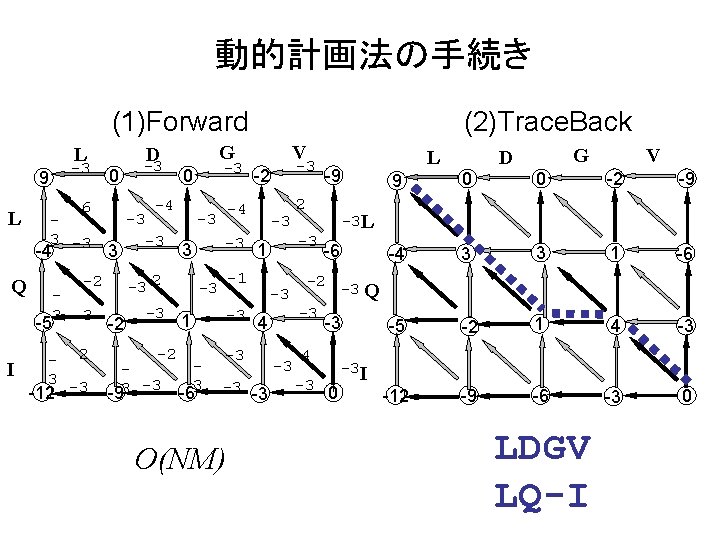
動的計画法の手続き (1)Forward L -3 9 L 6 3 -3 -4 3 -5 3 -3 - 2 3 -12 -3 -4 -3 -3 -2 -93 -3 -3 0 -3 -2 -4 -3 -3 -1 -3 -3 1 3 -6 -3 4 -3 -3 -3 O(NM) L -9 2 -3 -3 1 3 -3 2 -2 Q I 0 V G D -3 (2)Trace. Back -6 -2 -3 Q -3 4 -3 9 0 0 -2 -9 -4 3 3 1 -6 -5 -2 1 4 -3 -12 -9 -6 -3 0 -3 L -3 -3 V G D 0 -3 I LDGV LQ-I







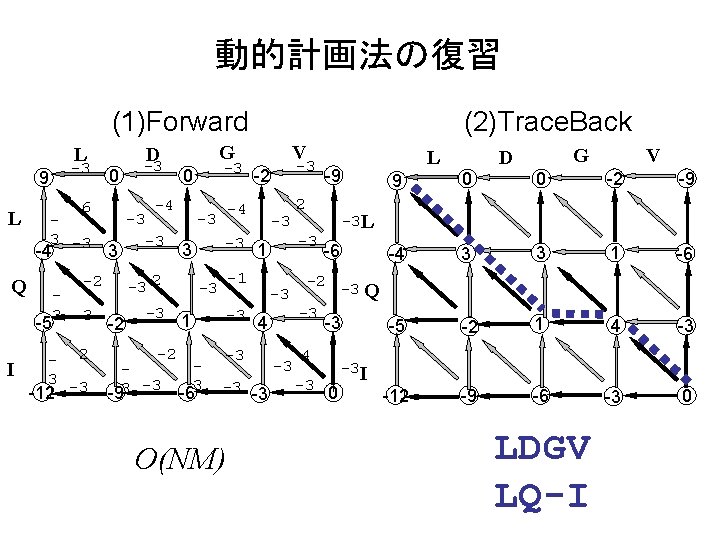
動的計画法の復習 (1)Forward L -3 9 L 6 3 -3 -4 3 -5 3 -3 - 2 3 -12 -3 -4 -3 -3 -2 -93 -3 -3 0 -3 -2 -4 -3 -3 -1 -3 -3 1 3 -6 -3 4 -3 -3 -3 O(NM) L -9 2 -3 -3 1 3 -3 2 -2 Q I 0 V G D -3 (2)Trace. Back -6 -2 -3 Q -3 4 -3 9 0 0 -2 -9 -4 3 3 1 -6 -5 -2 1 4 -3 -12 -9 -6 -3 0 -3 L -3 -3 V G D 0 -3 I LDGV LQ-I

![BLASTP 2 2 1 Apr13 2001 BLASTの 出力例1 Reference Altschul Stephen F Thomas BLASTP 2. 2. 1 [Apr-13 -2001] BLASTの 出力例(1) Reference: Altschul, Stephen F. , Thomas](https://slidetodoc.com/presentation_image_h/14e21021e916e7c1c34a6bdf78c8a5c7/image-44.jpg)
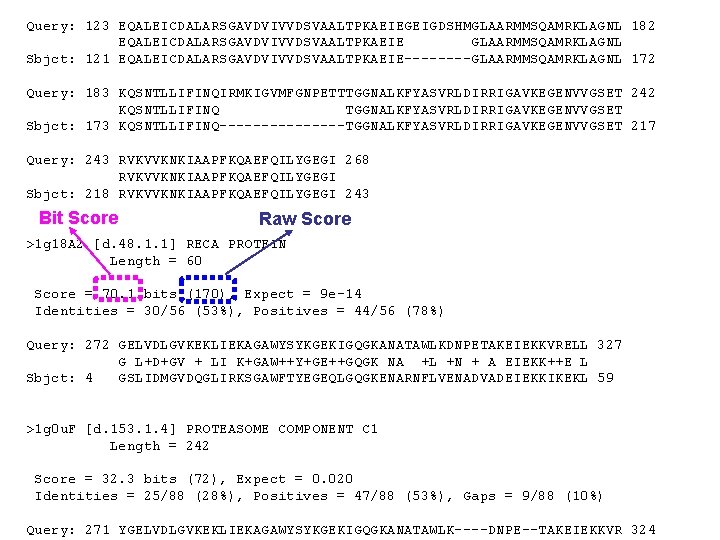
BLASTP 2. 2. 1 [Apr-13 -2001] BLASTの 出力例(1) Reference: Altschul, Stephen F. , Thomas L. Madden, Alejandro A. Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25: 3389 -3402. Query= RECA_ECOLI "Rec. A protein (Recombinase A)" (352 letters) Database: 40 scop 1. 59 nm 3886 sequences; 705, 110 total letters Searching. . . . done Score (bits) Sequences producing significant alignments: 2 reb-1 [c. 37. 1. 11] RECA PROTEIN (E. C. 3. 4. 99. 37) 1 g 18 A 2 [d. 48. 1. 1] RECA PROTEIN 1 g 0 u. F [d. 153. 1. 4] PROTEASOME COMPONENT C 1 1 byr. A [d. 136. 1. 1] ENDONUCLEASE 1 g 3 q. A [c. 37. 1. 10] CELL DIVISION INHIBITOR 1 ct 5 A [c. 1. 6. 2] YEAST HYPOTHETICAL PROTEIN, SELENOMET 1 g 0 u. D [d. 153. 1. 4] PROTEASOME COMPONENT PUP 2 1 e 32 A 2 [c. 37. 1. 13] P 97 1 g 0 u. A [d. 153. 1. 4] PROTEASOME COMPONENT Y 7 1 cp 2 A [c. 37. 1. 10] NITROGENASE IRON PROTEIN 1 f 3 o. A [c. 37. 1. 12] HYPOTHETICAL ABC TRANSPORTER ATP-BINDING PROTEIN 1 qj 2 B 2 [d. 133. 1. 1] CARBON MONOXIDE DEHYDROGENASE 1 dgy. A [c. 72. 1. 1] ADENOSINE KINASE 1 sky. B 3 [c. 37. 1. 11] F 1 -ATPASE 1 g 6 o. A [c. 37. 1. 13] CAG-ALPHA 1 cmx. A [d. 3. 1. 6] UBIQUITIN YUH 1 -UBAL 8 abp- [c. 93. 1. 1] L-*ARABINOSE-BINDING PROTEIN (MUTANT WITH MET 1. . . 2 tps. A [c. 1. 3. 1] THIAMIN PHOSPHATE SYNTHASE 448 70 32 28 28 28 27 26 26 26 25 25 25 24 24 24 E Value e-127 9 e-14 0. 020 0. 29 0. 38 0. 49 1. 1 1. 4 1. 9 2. 4 3. 2 4. 2 7. 1
![1 b 15 A c 2 1 2 ALCOHOL DEHYDROGENASE 1 pmi b 82 1 b 15 A [c. 2. 1. 2] ALCOHOL DEHYDROGENASE 1 pmi- [b. 82.](https://slidetodoc.com/presentation_image_h/14e21021e916e7c1c34a6bdf78c8a5c7/image-45.jpg)
1 b 15 A [c. 2. 1. 2] ALCOHOL DEHYDROGENASE 1 pmi- [b. 82. 1. 3] PHOSPHOMANNOSE ISOMERASE 23 23 >2 reb-1 [c. 37. 1. 11] RECA PROTEIN (E. C. 3. 4. 99. 37) Length = 243 9. 3 Score = 448 bits (1152), Expect = e-127 Identities = 243/266 (91%), Positives = 243/266 (91%), Gaps = 23/266 (8%) Query: 3 Sbjct: 1 Query: 63 Sbjct: 61 DENKQKALAAALGQIEKQFGKGSIMRLGEDRSMDVETISTGSLSLDIALGAGGLPMGRIV 62 DENKQKALAAALGQIEKQFGKGSIMRLGEDRSMDVETISTGSLSLDIALGAGGLPMGRIV 60 EIYGPESSGKTTLTLQVIAAAQREGKTCAFIDAEHALDPIYARKLGVDIDNLLCSQPDTG 122 EIYGPESSGKTTLTLQVIAAAQREGKTCAFIDAEHALDPIYARKLGVDIDNLLCSQPDTG 120 Query: 123 EQALEICDALARSGAVDVIVVDSVAALTPKAEIEGEIGDSHMGLAARMMSQAMRKLAGNL 182 EQALEICDALARSGAVDVIVVDSVAALTPKAEIE GLAARMMSQAMRKLAGNL Sbjct: 121 EQALEICDALARSGAVDVIVVDSVAALTPKAEIE----GLAARMMSQAMRKLAGNL 172 Query: 183 KQSNTLLIFINQIRMKIGVMFGNPETTTGGNALKFYASVRLDIRRIGAVKEGENVVGSET 242 KQSNTLLIFINQ TGGNALKFYASVRLDIRRIGAVKEGENVVGSET Sbjct: 173 KQSNTLLIFINQ--------TGGNALKFYASVRLDIRRIGAVKEGENVVGSET 217 Query: 243 RVKVVKNKIAAPFKQAEFQILYGEGI 268 RVKVVKNKIAAPFKQAEFQILYGEGI Sbjct: 218 RVKVVKNKIAAPFKQAEFQILYGEGI 243 >1 g 18 A 2 [d. 48. 1. 1] RECA PROTEIN Length = 60 Score = 70. 1 bits (170), Expect = 9 e-14 Identities = 30/56 (53%), Positives = 44/56 (78%) Query: 272 GELVDLGVKEKLIEKAGAWYSYKGEKIGQGKANATAWLKDNPETAKEIEKKVRELL 327 G L+D+GV + LI K+GAW++Y+GE++GQGK NA +L +N + A EIEKK++E L BLASTの 出力例(2)

Query: 243 RVKVVKNKIAAPFKQAEFQILYGEGI 268 RVKVVKNKIAAPFKQAEFQILYGEGI Sbjct: 218 RVKVVKNKIAAPFKQAEFQILYGEGI 243 >1 g 18 A 2 [d. 48. 1. 1] RECA PROTEIN Length = 60 Score = 70. 1 bits (170), Expect = 9 e-14 Identities = 30/56 (53%), Positives = 44/56 (78%) Query: 272 GELVDLGVKEKLIEKAGAWYSYKGEKIGQGKANATAWLKDNPETAKEIEKKVRELL 327 G L+D+GV + LI K+GAW++Y+GE++GQGK NA +L +N + A EIEKK++E L Sbjct: 4 GSLIDMGVDQGLIRKSGAWFTYEGEQLGQGKENARNFLVENADVADEIEKKIKEKL 59 >1 g 0 u. F [d. 153. 1. 4] PROTEASOME COMPONENT C 1 Length = 242 Score = 32. 3 bits (72), Expect = 0. 020 Identities = 25/88 (28%), Positives = 47/88 (53%), Gaps = 9/88 (10%) Query: 271 YGELVDLGVKEKLIEKAGAWYSYKGEKIGQGKANATAWLK----DNPE--TAKEIEKKVR 324 +G + G ++E +G+++ YKG G+G+ +A A L+ +PE +A+E K+ Sbjct: 132 FGGVDKNGAHLYMLEPSGSYWGYKGAATGKGRQSAKAELEKLVDHHPEGLSAREAVKQAA 191 Query: 325 EL--LLSNPNSTPDFSVDDSE-GVAETN 349 ++ L N DF ++ S ++ETN Sbjct: 192 KIIYLAHEDNKEKDFELEISWCSLSETN 219 >1 byr. A [d. 136. 1. 1] ENDONUCLEASE Length = 152 Score = 28. 5 bits (62), Expect = 0. 29 Identities = 28/102 (27%), Positives = 46/102 (44%), Gaps = 19/102 (18%) BLASTの 出力例(3)



![BLASTP 2 2 1 Apr13 2001 Reference Altschul Stephen F Thomas L Madden BLASTP 2. 2. 1 [Apr-13 -2001] Reference: Altschul, Stephen F. , Thomas L. Madden,](https://slidetodoc.com/presentation_image_h/14e21021e916e7c1c34a6bdf78c8a5c7/image-50.jpg)
BLASTP 2. 2. 1 [Apr-13 -2001] Reference: Altschul, Stephen F. , Thomas L. Madden, Alejandro A. Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25: 3389 -3402. Query= RECA_ECOLI "Rec. A protein (Recombinase A)" (352 letters) Database: 40 scop 1. 59 nm 3886 sequences; 705, 110 total letters Searching. . . . done Sequences producing significant alignments: 2 reb-1 [c. 37. 1. 11] RECA PROTEIN (E. C. 3. 4. 99. 37) 1 g 18 A 2 [d. 48. 1. 1] RECA PROTEIN 1 g 0 u. F [d. 153. 1. 4] PROTEASOME COMPONENT C 1 1 byr. A [d. 136. 1. 1] ENDONUCLEASE 1 g 3 q. A [c. 37. 1. 10] CELL DIVISION INHIBITOR 1 ct 5 A [c. 1. 6. 2] YEAST HYPOTHETICAL PROTEIN, SELENOMET 1 g 0 u. D [d. 153. 1. 4] PROTEASOME COMPONENT PUP 2 1 e 32 A 2 [c. 37. 1. 13] P 97 1 g 0 u. A [d. 153. 1. 4] PROTEASOME COMPONENT Y 7 1 cp 2 A [c. 37. 1. 10] NITROGENASE IRON PROTEIN 1 f 3 o. A [c. 37. 1. 12] HYPOTHETICAL ABC TRANSPORTER ATP-BINDING PROTEIN 1 qj 2 B 2 [d. 133. 1. 1] CARBON MONOXIDE DEHYDROGENASE 1 dgy. A [c. 72. 1. 1] ADENOSINE KINASE Score (bits) E Value 448 70 32 28 28 28 27 26 26 26 25 25 25 e-127 9 e-14 0. 020 0. 29 0. 38 0. 49 1. 1 1. 4 1. 9 2. 4 3. 2

Query: 123 EQALEICDALARSGAVDVIVVDSVAALTPKAEIEGEIGDSHMGLAARMMSQAMRKLAGNL 182 EQALEICDALARSGAVDVIVVDSVAALTPKAEIE GLAARMMSQAMRKLAGNL Sbjct: 121 EQALEICDALARSGAVDVIVVDSVAALTPKAEIE----GLAARMMSQAMRKLAGNL 172 Query: 183 KQSNTLLIFINQIRMKIGVMFGNPETTTGGNALKFYASVRLDIRRIGAVKEGENVVGSET 242 KQSNTLLIFINQ TGGNALKFYASVRLDIRRIGAVKEGENVVGSET Sbjct: 173 KQSNTLLIFINQ--------TGGNALKFYASVRLDIRRIGAVKEGENVVGSET 217 Query: 243 RVKVVKNKIAAPFKQAEFQILYGEGI 268 RVKVVKNKIAAPFKQAEFQILYGEGI Sbjct: 218 RVKVVKNKIAAPFKQAEFQILYGEGI 243 Bit Score Raw Score >1 g 18 A 2 [d. 48. 1. 1] RECA PROTEIN Length = 60 Score = 70. 1 bits (170), Expect = 9 e-14 Identities = 30/56 (53%), Positives = 44/56 (78%) Query: 272 GELVDLGVKEKLIEKAGAWYSYKGEKIGQGKANATAWLKDNPETAKEIEKKVRELL 327 G L+D+GV + LI K+GAW++Y+GE++GQGK NA +L +N + A EIEKK++E L Sbjct: 4 GSLIDMGVDQGLIRKSGAWFTYEGEQLGQGKENARNFLVENADVADEIEKKIKEKL 59 >1 g 0 u. F [d. 153. 1. 4] PROTEASOME COMPONENT C 1 Length = 242 Score = 32. 3 bits (72), Expect = 0. 020 Identities = 25/88 (28%), Positives = 47/88 (53%), Gaps = 9/88 (10%) Query: 271 YGELVDLGVKEKLIEKAGAWYSYKGEKIGQGKANATAWLK----DNPE--TAKEIEKKVR 324

Database: 40 scop 1. 59 nm Posted date: Jun 22, 2002 3: 06 PM Number of letters in database: 705, 110 Number of sequences in database: 3886 Lambda K H 0. 314 0. 134 0. 369 Gapped Lambda K H 0. 267 0. 0410 0. 140 Matrix: BLOSUM 62 Gap Penalties: Existence: 11, Extension: 1 Number of Hits to DB: 469, 543 Number of Sequences: 3886 Number of extensions: 18494 Number of successful extensions: 65 Number of sequences better than 10. 0: 17 Number of HSP's better than 10. 0 without gapping: 13 Number of HSP's successfully gapped in prelim test: 4 Number of HSP's that attempted gapping in prelim test: 50 Number of HSP's gapped (non-prelim): 17 length of query: 352 length of database: 705, 110 effective HSP length: 79 effective length of query: 273 effective length of database: 398, 116 effective search space: 108685668 effective search space used: 108685668

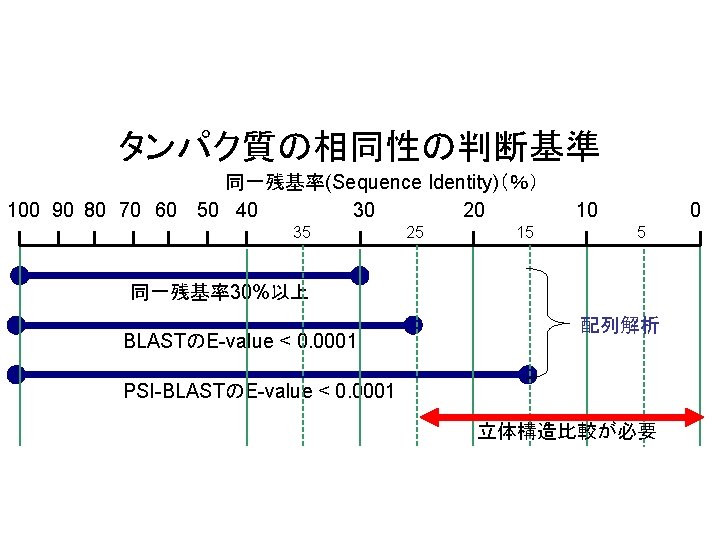
タンパク質の相同性の判断基準 100 90 80 70 60 同一残基率(Sequence Identity)(%) 50 40 30 20 35 25 15 10 0 5 同一残基率30%以上 BLASTのE-value < 0. 0001 配列解析 PSI-BLASTのE-value < 0. 0001 立体構造比較が必要


![blastpアミノ酸対アミノ酸によるタンパク質の機能予測 クエリT thermophiusのタンパク質 データベース大腸菌の全タンパク質 BLASTP 2 2 3 May13 2002 Query X 07 AAS blastp(アミノ酸対アミノ酸)によるタンパク質の機能予測 クエリ:T. thermophiusのタンパク質, データベース:大腸菌の全タンパク質 BLASTP 2. 2. 3 [May-13 -2002] Query= X 07 AAS](https://slidetodoc.com/presentation_image_h/14e21021e916e7c1c34a6bdf78c8a5c7/image-56.jpg)
blastp(アミノ酸対アミノ酸)によるタンパク質の機能予測 クエリ:T. thermophiusのタンパク質, データベース:大腸菌の全タンパク質 BLASTP 2. 2. 3 [May-13 -2002] Query= X 07 AAS 80531. 1 tthe 0 Database: ecoli_aa (144 letters) 4237 sequences; 1, 350, 094 total letters Sequences producing significant alignments: Score E (bits) Value inf. C NP_416233. 1 "protein chain initiation factor IF-3" NC_000913 rhs. D NP_415030. 1 "Rhs. D protein in Rhs. D element" NC_000913 pta NP_416800. 1 "phosphotransacetylase" NC_000913 prs. A NP_415725. 1 "phosphoribosylpyrophosphate synthetase" NC_000913 yia. K NP_418032. 1 "2, 3 -diketo-L-gulonate dehydrogenase, NADH-depe. . . ffh NP_417101. 1 "4. 5 S-RNP protein, GTP-binding export factor, pa. . . ybd. R NP_415141. 1 "putative dehydrogenase, NAD(P)-binding" NC_000913 ydf. G NP_416057. 1 "putative oxidoreductase" NC_000913 137 28 25 25 24 24 24 23 >inf. C NP_416233. 1 "protein chain initiation factor IF-3" NC_000913 Length = 180 Score = 137 bits (346), Expect = 2 e-34 Identities = 72/139 (51%), Positives = 92/139 (65%), Gaps = 1/139 (0%) Query: 4 Sbjct: 40 Query: 64 REALRLAQEMDLDLVLVGPNADPPVARIMDYSKWRYEQQMXXXXXXTEVKSIKFR 63 REAL A+E +DLV + PNA+PPV RIMDY K+ YE+ +VK IKFR REALEKAEEAGVDLVEISPNAEPPVCRIMDYGKFLYEKSKSSKEQKKKQKVIQVKEIKFR 99 VKIDEHDYQTKLGHIKRFLQEGHKVKVTIMFRGREVAHPELGERILNRVTEDLKDLAVVE 123 DE DYQ KL + RFL+EG K K+T+ FRGRE+AH ++G +LNRV +DL++LAVVE Sbjct: 100 PGTDEGDYQVKLRSLIRFLEEGDKAKITLRFRGREMAHQQIGMEVLNRVKDDLQELAVVE 159 2 e-34 0. 19 2. 0 2. 7 3. 5 4. 6 7. 8

blastp(アミノ酸対アミノ酸)の適用例) ORFのアノテーション: H. influenzaeのORF対大腸菌のORF Query= HI 0078 hinf 0 AAC 21753. 1 Sequences producing significant alignments: cys. S met. G ile. S leu. S yid. W ecol 0 ecol 0 AAC 73628. 1 AAC 75175. 1 AAC 73137. 1 AAC 73743. 1 AAC 76718. 1 "cysteine t. RNA synthetase" "methionine t. RNA synthetase" "isoleucine t. RNA synthetase" "regulator protein for dgo operon" Score E (bits) Value 730 39 39 30 28 0. 0 5 e-04 0. 001 0. 25 1. 3 → HI 0078はcysteine t. RNA syntetase Query= HI 0083 hinf 0 AAC 21762. 1 (71 letters) Sequences producing significant alignments: isp. B ecol 0 AAC 76219. 1 "octaprenyl diphosphate synthase" lpl. A ecol 0 AAC 77339. 1 "lipoate-protein ligase A" nlp. A ecol 0 AAC 76684. 1 "lipoprotein-28" b 1372 ecol 0 AAC 74454. 1 "putative membrane protein" mda. A ecol 0 AAC 73938. 1 "modulator of drug activity A" → HI 0083は大腸菌にはホモログがない Score E (bits) Value 23 22 22 3. 1 6. 9 9. 0
