Taverna Workflows my Experiment Paul Fisher University of





















































- Slides: 71

Taverna Workflows my. Experiment Paul Fisher University of Manchester http: //www. cs. man. ac. uk/~fisherp/Newcastle. Tutorial. ppt

Taverna This tutorial is designed to introduce you to the Taverna 1. 7 workflow workbench

1. Installing the Workbench

Prerequisites - 1 Java In order to run Taverna 1. 7 on your computer you will need to have the latest Java installed. If you do not have Java already installed, you can download it from this URL: http: //java. sun. com/javase/downloads/index. jsp You will have a choice of the download you would like. Download the JDK with Java EE packaged up too. This will give you the opportunity to develop web services and use the ones deployed by Java developers at a later date. The Java Runtime Environment (JRE) being downloaded should be 1. 5 or later for Taverna to work. If you have Java installed, but it is an earlier version, you will need to update it to 1. 5 or later otherwise Taverna will NOT work. The minimal installation you will need is the standard JDK package. Download the desired JDK by following the link on the website and choose a location on your computer to save it to. Open the saved file and follow the installation instructions to install Java on your computer Restart your computer to complete the installation.

Prerequisites - 2 A zip package You will also need a tool to unzip the downloaded workbench. There are various tools available on the internet, including Win. Zip, 7 -Zip, and a few others. Personally I prefer 7 Zip, which is free to and easy use, available at the following URL: http: //www. 7 -zip. org/download. html You will need to choose the appropriate file to download for your operating system, i. e. Windows, Linux, Apple MAC. Choose a location to save the file in and save it. Locate your saved file and follow the installation instructions to install it on your computer. Restart your computer to complete the installation.

Prerequisites - 3 Linux users - Graphviz Those who are installing Taverna on Linux will also have to install Graphviz onto the system. This is available at the following URL: http: //www. graphviz. org/ At the time of writing – I have no installation instructions for this package, so please refer to the user documentation provided on the web site

Downloading Taverna Open your usual web browser and go to the my. Grid homepage at the following URL: http: //www. mygrid. org. uk Find and follow the links to download Taverna 1 link on the web page http: //www. mygrid. org. uk/tools/taverna-1/taverna-download/ Once on the ‘Download’ page, identify the relevant Taverna distribution you need. Follow the link to download the workbench. The web page should re-direct you to the source forge page. Choose a location to save the file and click OK.

Unzipping the workbench Choose to “Unzip/Extract the files”, but not into the current directory. You will need to choose a directory in which to unzip the files. I recommend somewhere in the root drive of your computer so you can easily access it, e. g. C: my. Grid. You can change the name of the folder at this stage, e. g. to “Taverna”. If you are using Taverna on Linux, please be sure that you have the relevant access permissions to install and run Taverna in the desired directory. If you need a Zip package – download and install “ 7 -ZIP” (find it using Google)

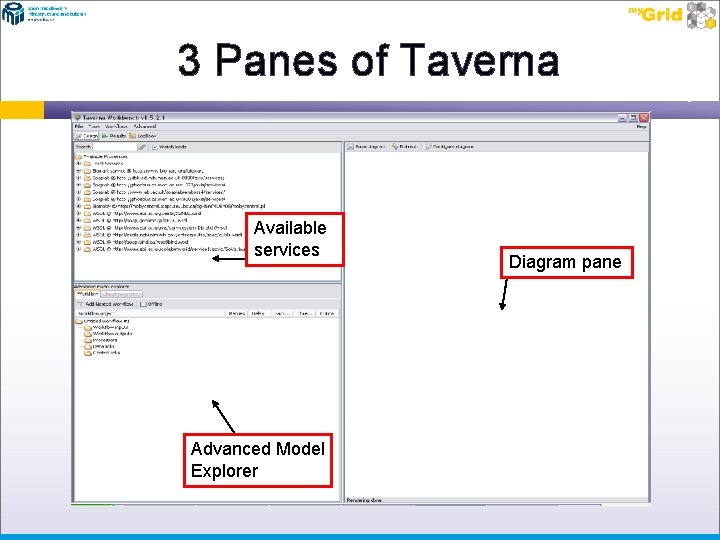
Opening Taverna Locate you Taverna installation and open the Taverna folder. Start Taverna by double clicking on the “runme. bat” (Windows users )or “runme. sh” (Linux and Mac users). If you have successfully installed Java, you should see a dialog box or command window open, shortly followed by the Taverna application. Once you have installed Taverna for the first time it will need to update all of its components. You do not need to do anything for this, as this happens as the workbench is opening. You should see a graphic in the centre of your screen, with a download progress. Each component will be shown loading in this progress bar in turn. Once this has completed (depending on connection speed – about 5 minutes), the Taverna workbench will open. The Taverna workbench consists of 3 main panels for constructing workflows: � � � The Available services pane (Top Left side) The Advanced Model Explorer pane (Bottom Left side) The Diagram pane (Right side)

The 3 Panes of Taverna The Available services pane is used to display the web services to the user. This list contains default services from when the workbench starts. Once you become more experienced with the workbench, you will be able to add you own services, including adding default services so they load automatically when Taverna opens. This list contains WSDL web services, local Bio. Java widgets, Soaplab services, and Bio. Moby objects. Each of these can be added to the workflow model (workflow being constructed) so that a task can be achieved. The Advanced Model Explorer (AME) pane contains the services used in the current workflow, including the inputs, outputs, and data links between each service. Once populated with services, each service can be expanded using the “+” button. This provides a list of the inputs and outputs that the service takes in and expels. It is these inputs and outputs that allow you to connect services together. The Diagram pane shows a graphical representation of the workflow being used/constructed. The diagram can be adapted to view different aspects of the current workflow, to show all the ports for all the services, only those ports that have been connected or bound, or to change the layout of the workflow from portrait to landscape.

3 Panes of Taverna Available services Advanced Model Explorer Diagram pane

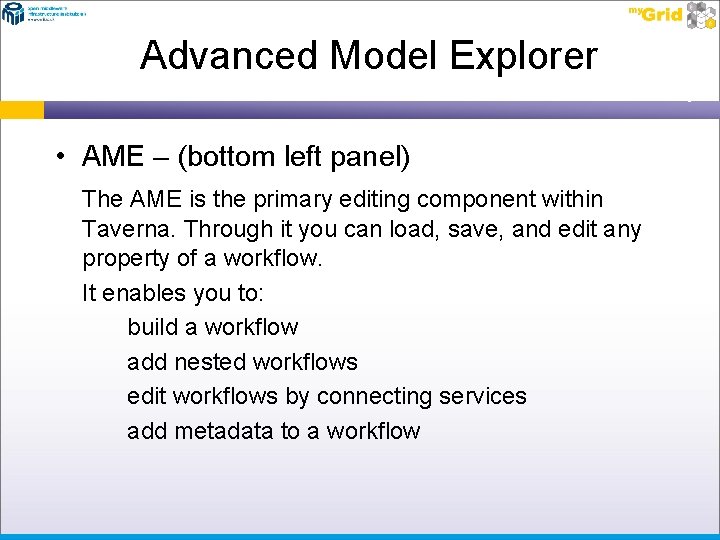
Advanced Model Explorer • AME – (bottom left panel) The AME is the primary editing component within Taverna. Through it you can load, save, and edit any property of a workflow. It enables you to: build a workflow add nested workflows edit workflows by connecting services add metadata to a workflow

Diagram Pane • Shows inputs / outputs, services and control flows • It allows you to change the view of a workflow, save the visual representation, and explode or implode nested workflows

Available services Lists services available by default in Taverna – top left • ~ 3500 services Local java services Simple web services Soaplab services – legacy command-line application R Processor Bio. Mart database services Bio. Moby services Beanshell processor Allows the user to add new services or workflows from the web or from file systems

2. Adding new services

Exercise 2 Adding New Services New services can be gathered from anywhere on the web – the default list are just a few we already know about – importing others is very straightforward � Go to the DDBJ list of available web services at: http: //xml. nig. ac. jp/wsdl/index. jsp These services were not designed for use in Taverna, but Taverna can use them if you supply the address of the WSDL file Click on the DDBJ blast service (http: //xml. nig. ac. jp/wsdl/Blast. wsdl) and copy the web page address

Exercise 2 Adding New Services Go to the services panel in Taverna, and right-click on ‘Available Processors’ (at the top of the list). For each type of service, you are given the option to add a new service, or set of services. Select ‘Add new WSDL scavenger’. A window will pop-up asking for a web address Enter the Blast Web service address you just copied Scroll down to the bottom of the Services list and look at the new DDBJ service that is now included, clicking on the “+” icon next to the service

3. Finding and Invoking a Service

Exercise 3 - Finding and invoking a Service Go to the Services Panel Type ‘binfo’ into the search box at the top of the panel (we will start with simple information retrieval from KEGG) You may see several services highlighted in red Scroll down to the KEGG services, to ‘binfo’ This service returns information about the KEGG databases, depending on the information you supply to it, e. g. the word ‘pathway’ gives info on the KEGG pathway database

Exercise 3 Invoking a single service Right click on the ‘binfo’ service and select ‘Invoke service’ In the pop-up ‘Run workflow’ window add the word “pathway” by clicking on the input document ‘db’ and selecting to ‘add new input’ from the dialog menu. Click ‘Run workflow’ and the service is invoked

Exercise 3 View Results Click on the ‘Results’ tab in the Taverna tool bar The database information is displayed on the right when you select ‘click to view’ Click on the ‘Process Report’ tab Look at processes. This shows the experimental provenance – where and when processes were run, and times Click on the ‘Status’ tab Look at options As workflows run, you can monitor their progress here (Note: this workflow was probably too fast to see this feature properly, we will come back to it later)

Exercise 3 - Conclusion The processes for running and invoking a single service are the basics for any workflow and the tracking of processes and generation of results are the same however complicated a workflow becomes In the next few exercises, we will look at some example workflows and build some of our own from scratch

4. Finding and Using Workflows

Installing The Whip Plug-in Your going to use the ‘new’ my. Experiment Plug-in Firstly you need to install WHIP - http: //www. whipplugin. org/ This allows you to interact with the my. Experiment server In Taverna, go to “Tools” and then select “Plug-in Manager” Click “Find New Plug-ins”, and select the “my. Experiment and WHIP (beta) plug-in” from the list Then click “Install” to install the plug-in

You should now see the my. Experiment plug-in in the toolbar menu Browse through the example workflows in the first tab of the plug-in To view a workflow, select “Preview” from the buttons under the workflow diagram To open a workflow in the workbench, click on the open button under the workflow diagram

Previewing a workflow allows you to see all the metadata associated with the workflow on the my. Experiment website, including: TAGS � AUTHOR � CREDITS � DESCRIPTION � You can also view the latest workflows, search for keywords, and even browse using a tag cloud � Choose a workflow to load and click on “Open” �

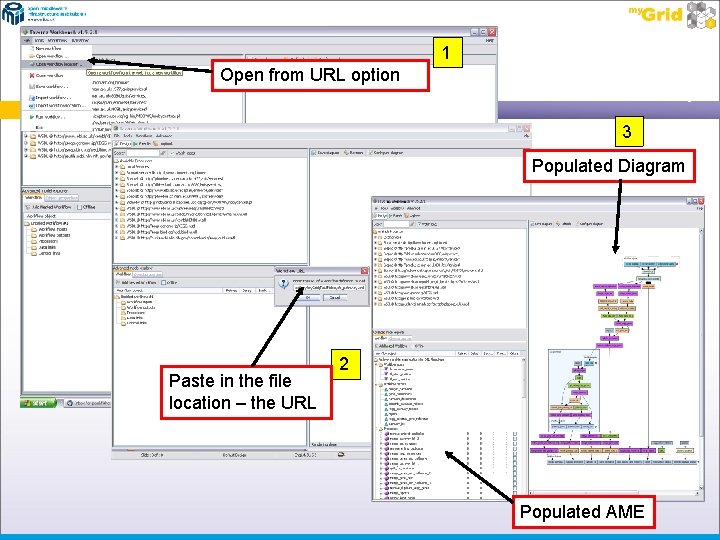
Opening from a URL Select ‘Open Workflow Location’ from the File menu at the top of the workbench. In the pop-up window, add the following web address to load a workflow from the web http: //www. myexperiment. org/workflows/16/download? version=3 The ‘Mouse Pathways and Gene annotations for QTL Phenotype’ workflow will be loaded View the workflow diagram - you will see services in a couple of different colours

1 Open from URL option 3 Populated Diagram Paste in the file location – the URL 2 Populated AME

Exercise 4 Workflow Documentation In the Advanced Model explorer panel – click on the name of the workflow at the top of the window (just above Inputs) – in this case ‘Pathways and Gene annotations for QTL Phenotype’ and then select the ‘workflow metadata’ tab at the top of the AME. You will see a text description of the workflow, its author and its unique LSID (Life Science Identifier). When publishing workflows for others, this annotation is useful information and allows the acknowledgement of intellectual property

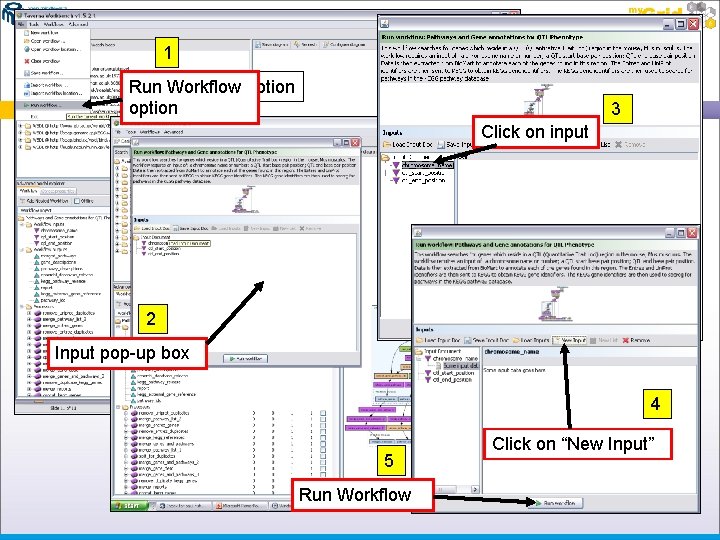
Exercise 4 Workflow Features Now that you have loaded your workflow you can execute it To execute your workflow open the “File” Menu at the top of the Workbench Choose “Run Workflow” from the options given – this will open a pop-up box to input your data Each input requires you to enter data – to enter data into each of the inputs, click on one input and then click on the “New Data” option in the pop-up menu system Once you have entered these details, press the “Run Workflow” button at the bottom of the pop-up box

1 Run Workflow option 3 Click on input 2 Input pop-up box 4 5 Run Workflow Click on “New Input”

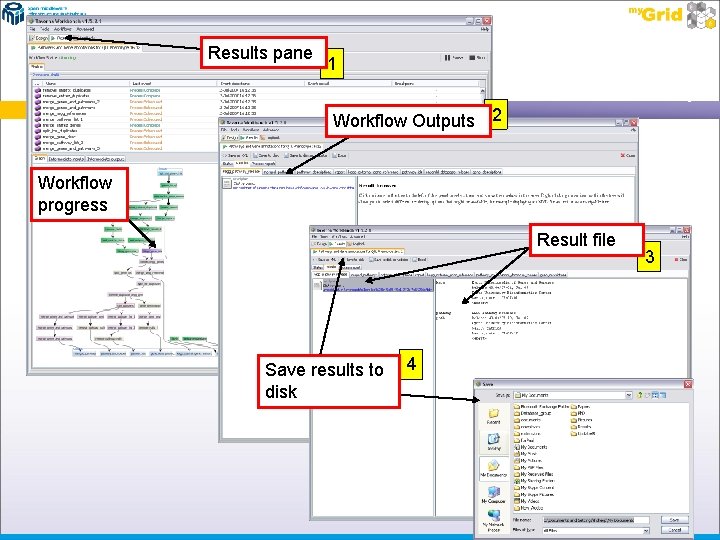
Viewing Results Once you have executed the workflow, the Taverna workbench will change views from “Design” to “Results”. You should see this change behind you Input pop-up box You can minimise the Input pop-up box to view the progress of the workflow being executed – the different colours indicate whether a service has run or not � Green = Completed � Purple = Currently being executed � Grey = Awaiting execution Once completed, the results will appear as separate tabs at the top of the workflow diagram (indicated in the following diagram as workflow outputs) Each tab contains an output file of results – the results can be viewed by clicking on the file in the left hand pane where it says “click to view” The file can then be searched through using the right hand pane, allowing you to verify the results – if they are wrong simply maximise the pop-up window and hit the “Run workflow” button again, making sure that the inputs are correct Each file can then be saved to the local machine – to do this simply click on the button marked “Save to disk” and enter the location to save the files Then click OK

Results pane 1 Workflow Outputs 2 Workflow progress Result file Save results to disk 4 3

5&6 Building a simple workflow

5. 1 Building a simple workflow from scratch Import the ‘get_genes_by_pathway’ service into a new workflow model. First, you will need to either close the current workflow from the file menu, or select ‘New Workflow’ then find the above service again in the ‘services’ search panel. Right-click on ‘get_genes_by_pathway’ and import it into the workbench by right clicking, and selecting ‘Add to Model’ Go to the AME and expand the [+] next to the newly imported service. You will see: 1 input (Green arrow pointing up) 1 output (purple arrow pointing down)

Exercise 5. 2 Adding Input Define a new workflow input by right-clicking on ‘Workflow Input’ and selecting ‘Create New Input’ Supply a suitable name e. g. ‘pathway_identifier’ Connect this new input to the ‘get_genes_by_pathway’ service by right-clicking on ‘pathway_identifier’ and selecting ‘get_genes_by_pathway ->pathway_id’ You always build workflows with the flow of data

Exercise 5. 3 Adding output Define a new workflow output by right-clicking on ‘workflow output’ and selecting ‘create new output’ Supply a suitable name e. g. ‘gene_outputs’ Connect the ‘get_genes_by_pathway’ service to the new output, remembering to build with the flow of data You have now built a simple workflow from scratch! Run the workflow by selecting ‘run workflow’ from the ‘File’ menu at the very top of the workbench. You will again need to supply a KEGG pathway identifier – “path: mmu 03010”

Exercise 6 String Constants Select a ‘string constant’ from ‘Available Services’ list (by searching for ‘constant’ in the text search box Right-click and select ‘add to model with name…’ Insert ‘pathway_id’ in the pop-up window In the AME, right-click on ‘pathway_id’ and select ‘edit me’ Edit the text to ‘path: mmu 03010’. Replace the workflow input with this string constant Run the workflow – it runs in the same way Add a description and your name as author to the metadata section Save the workflow by selecting ‘save’ in the file menu

Exercise 7 Defining Output Formats So far, most of the outputs we have seen have been text, but in bioinformatics, we often want to view a graph, a 3 D structure, an alignment etc. Taverna is able to display results using a specific type of renderer if the workflow output is configured correctly. Load the ‘Fetch PDB flatfile from RCSB server’ workflow from http: //www. myexperiment. org/workflows/167/download? version=1 Run the workflow with the ID ‘ 1 crn’, or another PDB id you know of

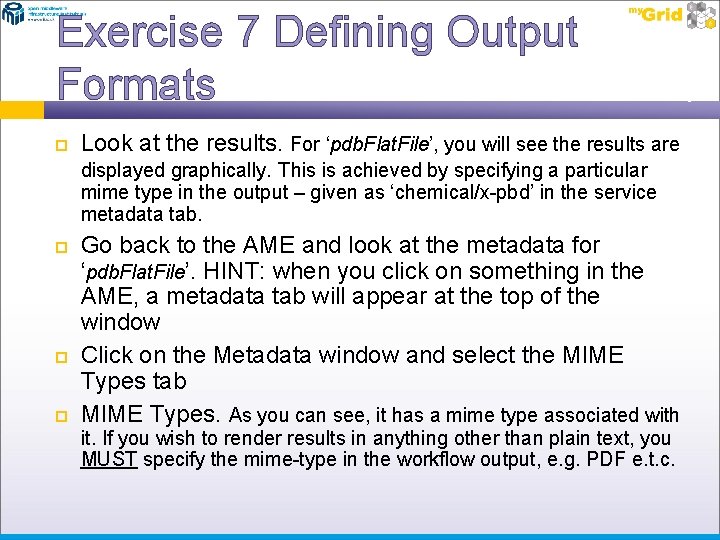
Exercise 7 Defining Output Formats Look at the results. For ‘pdb. Flat. File’, you will see the results are displayed graphically. This is achieved by specifying a particular mime type in the output – given as ‘chemical/x-pbd’ in the service metadata tab. Go back to the AME and look at the metadata for ‘pdb. Flat. File’. HINT: when you click on something in the AME, a metadata tab will appear at the top of the window Click on the Metadata window and select the MIME Types tab MIME Types. As you can see, it has a mime type associated with it. If you wish to render results in anything other than plain text, you MUST specify the mime-type in the workflow output, e. g. PDF e. t. c.

Exercise 7 Taverna MIMETypes The following mime-types are currently used by Taverna text/plain=Plain Text text/xml=XML Text text/html=HTML Text text/rtf=Rich Text Format text/x-graphviz=Graphviz Dot File image/png=PNG Image image/jpeg=JPEG Image image/gif=GIF Image application/zip=Zip File chemical/x-swissprot=SWISSPROT Flat File chemical/x-embl-dl-nucleotide=EMBL Flat File chemical/x-ppd=PPD File chemical/seq-aa-genpept=Genpept Protein chemical/seq-na-genbank=Genbank Nucleotide chemical/x-pdb=Protein Data Bank Flat File chemical/x-mdl-molfile

Exercise 8 Sharing Workflows Go to http: //www. myexperiment. org my. Experiment is a social networking site for sharing workflows and workflow expertise and experiences Browse around the site and see what it contains Create yourself an account and join the group called “Newcastle MSc. ” (this will be necessary for the next exercise)

Exercise 8 Sharing workflows Find all the workflows containing BLAST searches. How did you find them? How many are there? Can they all be downloaded? Which is the most downloaded workflow? Which is the most viewed workflow? Is it the same? How many workflows are tagged with ‘protein_structure’ ? If you wish to share your workflows with the rest of the class, upload them and set the permissions so that only those in the ‘Newcastle MSc. ’ group can see them – make sure you add a description and author details to the workflow metadata first!

Exercise 9 Workflow Reuse – Nested Workflows Reload your KEGG workflow from exercise 6 We will extend this workflow to get descriptions of each gene identifier, and find the pathways for each gene. In the my. Experiment plug-in, find all the workflow that are tagged with KEGG Select the ‘Get Kegg Gene information’ workflow http: //www. myexperiment. org/workflows/611

Exercise 9 Workflow Reuse – Nested Workflows Go back to Taverna and look at the original workflow In the AME, click on ‘add nested workflow’. Go back to the my. Experiment plug-in, and choose to “import from URL” for the workflow you found in my. Experiment You can change the name of the nested workflow by right-clicking on the processor and selecting ‘rename’, on the nested workflow You need to connect up the workflow as if it was any other kind of service

Exercise 9 Workflow Reuse – Nested Workflows The nested workflow has 1 input and 2 outputs. We have to connect the input, but we can choose which outputs to display In the outer workflow create a new output called ‘gene_descriptions’ - hint: to switch between workflows, use the “Workflows” option in the file menu system Connect gene_descriptions to the nested workflow output ‘gene_descriptions’

Exercise 9 Workflow Reuse – Nested Workflows Save the workflow (remembering to embed the nested workflow, using the supplied check box) and run the workflow Look at the results

Exercise 10 Iteration Taverna has an implicit iteration framework. If you connect a set of data objects (for example, a set of fasta sequences) to a process that expects a single data item at a time, the process will iterate over each sequence Load the ‘Mouse Pathways and Gene annotations for QTL Phenotype’ workflow from the my. Experiment plug-in using any of the previously used import methods http: //www. myexperiment. org/workflows/16/download? version=3 Watch the progress report. You will see several services with ‘Invoking with Iteration’

Exercise 10 Iteration � The user can also specify more complex iteration strategies using the service metadata tag Find and load the workflow ‘Demonstration of configurable iteration’ from the my. Experiment plug-in Read the workflow metadata to find out what the workflow does Select the ‘Colour. Animals’ service and read the metadata for that service. Under the description is the iteration strategy Click on ‘dot product’. This allows you to switch to cross product

Exercise 10 Iteration Run the workflow twice – once with ‘dot product’ and once with ‘cross product’. Save the first results so you can compare them – what is the difference? What does it mean to specify dot or cross product?

Exercise 11 Substituting Services Taverna does not own many of the bioinformatics services it provides. This means that it cannot control their reliability. Instead, Taverna provides strategies for dealing with services being unavailable Load the ‘Biomart. And. EMBOSSAnalysis’ from the my. Experiment website this time, using the ‘Launch in Taverna’ button. http: //www. myexperiment. org/workflows/158 Look at the metadata for the ‘emma’ service. It is an implementation of clustalw Find the DDBJ clustalw service – HINT: go to the DDBJ services homepage, and import the service from URL into the Available Services palatte http: //xml. nig. ac. jp/index. html

Exercise 11 Substituting Services Instead of adding the new service normally, right-click and select ‘add as alternate’ In the resulting menu select ‘emma’ The DDBJ version of the Clustal. W service is now added as an alternative to emma in the AME. It will appear at the bottom of the input/output list of the Emma service Select the new service (which should be called ‘analyze. Simple’ and look at the inputs and outputs. These need to be connected to the correct inputs and outputs in Emma (it is unlikely the inputs and outputs will have the same names! – see if you can figure them out)

Exercise 11 Substituting Services Right-click on the ‘query’ input in analyze. Simple and map it to ‘sequence_direct_data’. In both services, these inputs expect a set of fasta sequences. Right-click on the ‘result’ output and map it to ‘outseq’ in emma in the same way. Now you have a workflow which will run using emma when it is available – but will substitute it for DDBJ clustalw if emma fails!

Exercise 12 Failover Taverna also allows the user to specify the number of times a service is retried before it is considered to have failed. Sometimes network traffic is heavy, so a working service needs to be retried Select ‘tmap’ from the same workflow. To the right of the service name are a series of 0 s and 1 s. By simply typing the numbers, the user can specify the number of retries and the time between the retries Change it to 3 retries for ‘tmap’ and set the status to ‘critical’ using the final tickbox. Now it is critical, it means the whole workflow will be aborted if ‘tmap’ fails after 3 retries. Failures in non-critical services will not abort the workflow run.

Spotlight on Bio. Mart

Exercise 13 Spotlight on Bio. Mart Biomart enables the retrieval of large amounts of genomic data e. g. from Ensembl and Sanger, as well as Uniprot and MSD datasets After saving any workflows you want to keep, reset the workbench in the AME (by closing open workflows in the File menu) Keep open the workflow ‘Biomart. And. EMBOSSAnalysis’ Run the Workflow

Exercise 13 Spotlight on Bio. Mart This Workflow Starts by fetching all gene IDs from Ensembl corresponding to human genes on chromosome 22 implicated in known diseases and with homologous genes in rat and mouse. For each of these gene IDs it fetches the 200 bp after the five-prime end of the genomic sequence in each organism and performs a multiple alignment of the sequences using the EMBOSS tool 'emma' (a wrapper around Clustal. W). It then returns PNG images of the multiple alignment along with three columns containing the human, rat and mouse gene IDs used in each case.

Exercise 13 Spotlight on Bio. Mart Right-click on the ‘hsapiens_gene_ensembl’ service and select ‘configure Bio. Mart query’ By selecting ‘Filters’ and then ‘Region’ – change the chromosome from 22 to 21 – now the workflow will retrieve all disease genes from chromosome 21 with rat and mouse homologues Run the workflow and look at the results See how some of the other options were configured by finding them in the other pull-down lists (Gene, Multi-species comparison etc)

Exercise 13 Spotlight on Bio. Mart Find out which Gene Ontology terms are associated with the genes in your region by adding a new Biomart query processor Select another copy of ‘hsapiens_gene_ensembl’ from the services panel (under Biomart and Ensembl 50 genes (Sanger)) and select ‘add to model with name…. ’ (as there is already a service with that name!) and call the service ‘hsapiens_GO’ Configure ‘hsapiens_GO’ by right-clicking and selecting ‘configure Biomart query’ and selecting ‘filters’. In filters, select ‘gene’ and the ‘id list limit’ tick-box next to ‘ensembl gene IDs’. Configure the output (by selecting attributes) and select ‘GO ID’ for each GO partition under the ‘External -> GO Attributes’ tab in the attributes section

Exercise 13 Spotlight on Bio. Mart Connect the input to the ‘hsapiens_gene_ensembl’ service via the ‘ensembl_gene_id’ Create 3 new workflow outputs, ‘CCGOID’, ‘MFGOID’ and ‘BPGOID’. Connect the outputs of the biomart processor to them Re-run the workflow and view which GO terms are associated with your chromosomal region NOTE: Having 3 outputs for related terms like this is inefficient and hard to read – we will come back to a solution to fix this problem in the next session

Shim Services This exercise highlights the services that do not perform biological functions, but are vital for running life science workflows

Exercise 14 A shim is a service that doesn’t perform an experimental function, but acts as a connector, or glue when 2 experimental services have incompatible outputs and inputs A shim can be any type of service – WSDL, Soaplab etc. Many are simple Bean. Shell scripts

Exercise 14 – Finding Shims In the ‘Biomartand. Emboss. Analysis’, work out which services are shims What do the shims do?

Exercise 14 Other Shims There are many my. Grid shim services. These are currently being described in a shim library, but for now, a small collection are documented here: http: //www. cs. man. ac. uk/~hulld/shims. html Find a shim that will return a DNA file in Fasta format from an id. Load the example workflow and run it in Taverna Find a shim that will translate DNA HINT: these services might be in the feta registry

Exercise 14 Other Shims The emboss suite of programs have a subdivision – edit All the edit services are shims Experiment with the edit services Find a service that will remove gaps from sequences

Exercise 14 Beanshell Open Taverna and load the workflow ‘Biomart. And. EMBOSSAnalysis’ Look at the diagram. Each brown service is a Bean. Shell script In the ‘Advanced Model Explorer’ (AME) select the Bean. Shell ‘Create. Fasta’ Right-click and select ‘configure beanshell’

Exercise 14 Beanshell Look at the script and see if you can work out its function Look at the ports and their types as well as the script Note the names of the ports and where they appear in the script, you will need to know how to specify an input/output in the next exercise

Exercise 14 Beanshell – Writing your Own Beanshell scripts allow users to write small, bespoke java scripts to allow incompatible services to work together Create a new workflow by selecting ‘file’ and ‘New Workflow’ q q Add a new beanshell processor by right-clicking “Beanshell scripting host” in the service panel and selecting “Add to model” (you may change the name of the processor) Right click the beanshell processor created and select “ Configure beanshell…” q Create 2 input port named: my. Name and my. Surname q Cretate 1 output port named: my. Fullname Note that theses ports are automatically added to AME window

Exercise 14 Beanshell – Writing your Own Select the script tab and Paste the following script my. Fullname = my. Name +"t" + my. Surname q Create 2 workflow inputs and 1 workflow output by going to the port menu, and choosing to add a new port for both input and output. q Connect them to the configured beanshell processor. q Run the workflow q You should get your full name printed in the output q

Bio. Catalogue is a social networking site that allows you to discover Web Services, to include in your workflows Go to http: //www. biocatalogue. org Familiarise yourself with the page Go to ‘Project information’ and look at the roadmap to see what features are coming If you want to try Bio. Catalogue, you can sign up to the friends email list (found on the front page at the bottom left), and you can try the Pilot out by signing up for the beta testing: http: //beta. biocatalogue. org/ 1. 2. Username: biocat Password: biodog

FINISH
 Disease-specific workflows
Disease-specific workflows Jira default workflow diagram
Jira default workflow diagram Ivar nnn
Ivar nnn Goanywhere advanced workflows
Goanywhere advanced workflows Process discriminants in software project management
Process discriminants in software project management Sirsi workflows
Sirsi workflows Istituto comprensivo taverna di montalto
Istituto comprensivo taverna di montalto Noite na taverna personagens secundários
Noite na taverna personagens secundários Taverna workbench
Taverna workbench Taverna rovn
Taverna rovn Taverna workbench
Taverna workbench What is the main conflict in tangerine
What is the main conflict in tangerine Jean-paul rodrigue hofstra university
Jean-paul rodrigue hofstra university De paul university
De paul university Nyu avery fisher center
Nyu avery fisher center Derived properties of powder
Derived properties of powder Fisher-race nomenclature
Fisher-race nomenclature Simplified fisher equation
Simplified fisher equation Fisher exact test
Fisher exact test Indice di fisher formula
Indice di fisher formula Fisher transform indicator pdf
Fisher transform indicator pdf Kiss of the vampire a level media representation
Kiss of the vampire a level media representation Test exacto de fisher
Test exacto de fisher Fisher effect formula
Fisher effect formula Louis kahn fisher house
Louis kahn fisher house Công thức fisher
Công thức fisher The city of refuge rudolph fisher
The city of refuge rudolph fisher Costs of inflation
Costs of inflation Paasche weighted aggregate method
Paasche weighted aggregate method Fisher model
Fisher model Fisher 2390 level transmitter manual
Fisher 2390 level transmitter manual Thermo fisher core values
Thermo fisher core values Fisher exact test
Fisher exact test Fin221
Fin221 Tilly fisher
Tilly fisher Tipos de curtosis
Tipos de curtosis Test exacto de fisher
Test exacto de fisher Uses of index number
Uses of index number Clark fisher model uk
Clark fisher model uk Distribucion de fisher
Distribucion de fisher Fisher separation theorem assumptions
Fisher separation theorem assumptions Calculating purchasing power parity
Calculating purchasing power parity Função linear
Função linear Formula fisher
Formula fisher Fisher model
Fisher model Mr zeev duieb
Mr zeev duieb Thermo fisher scientific mission statement
Thermo fisher scientific mission statement Evelyn gerlach
Evelyn gerlach Fisher information exponential family
Fisher information exponential family Peter fisher physics
Peter fisher physics Charlotte fisher target
Charlotte fisher target Narrative paradigm
Narrative paradigm Walter fisher narrative paradigm
Walter fisher narrative paradigm Fisher family kinetic letters
Fisher family kinetic letters Fisher separation theorem graphically
Fisher separation theorem graphically Fisher árindex
Fisher árindex Theories of intellectual property william fisher
Theories of intellectual property william fisher Fisher féle volumenindex
Fisher féle volumenindex Innovative and functional products
Innovative and functional products Mun engineering complementary studies
Mun engineering complementary studies Distribusi f
Distribusi f Fischer effect formula
Fischer effect formula Fisher effect equation
Fisher effect equation Think said the robin think said the jay
Think said the robin think said the jay Enantiomeros
Enantiomeros Rita moreno flag
Rita moreno flag Farah fisher
Farah fisher Limitations of fisher's quantity theory of money
Limitations of fisher's quantity theory of money Jenis-jenis mechanical fastening
Jenis-jenis mechanical fastening Fisher science education
Fisher science education Fisher house west palm beach
Fisher house west palm beach What is the endowment point in economics
What is the endowment point in economics