Studio del trasporto citoplasmanucleo Trasporto dellm RNA Trasporto





- Slides: 30

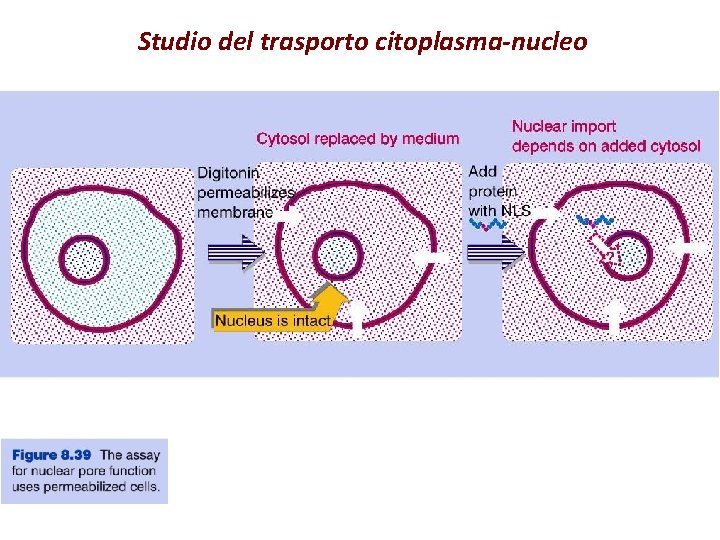
Studio del trasporto citoplasma-nucleo


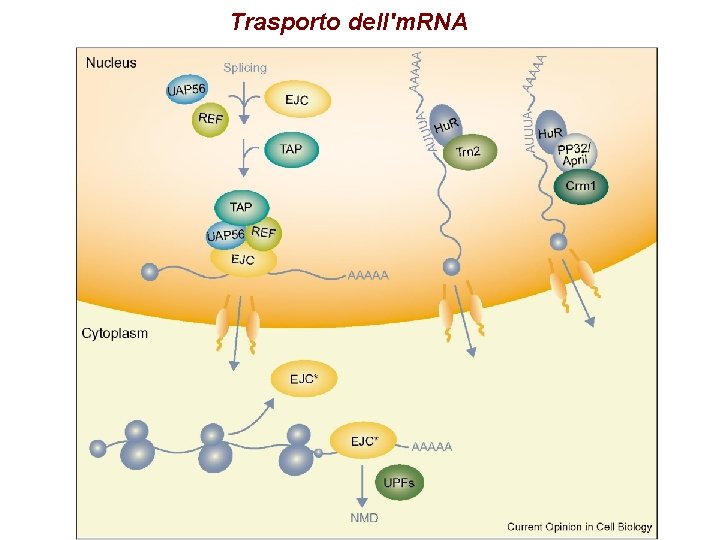
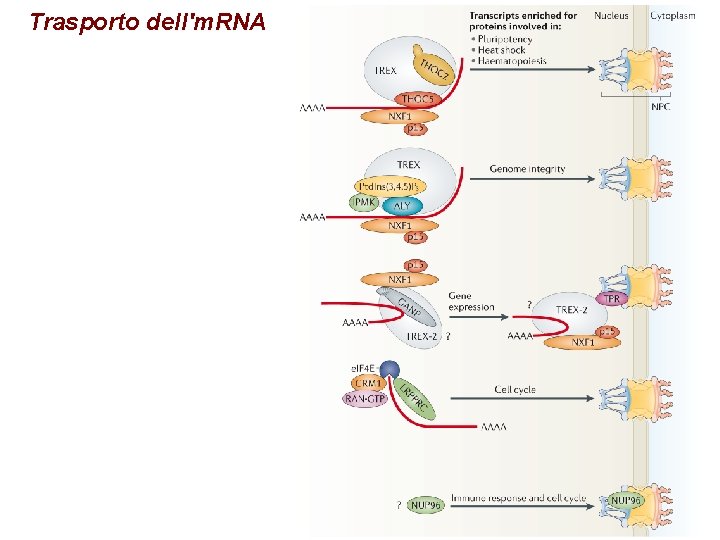
Trasporto dell'm. RNA

Trasporto dell'm. RNA

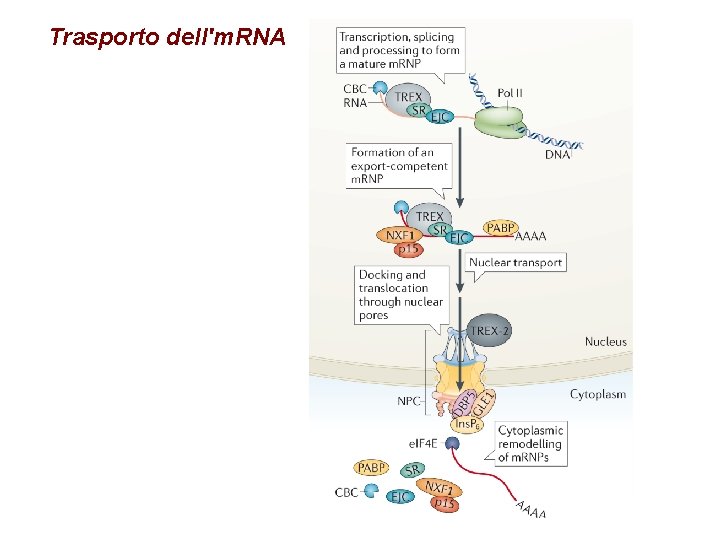
Trasporto dell'm. RNA





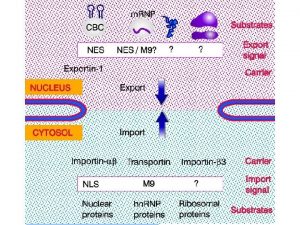
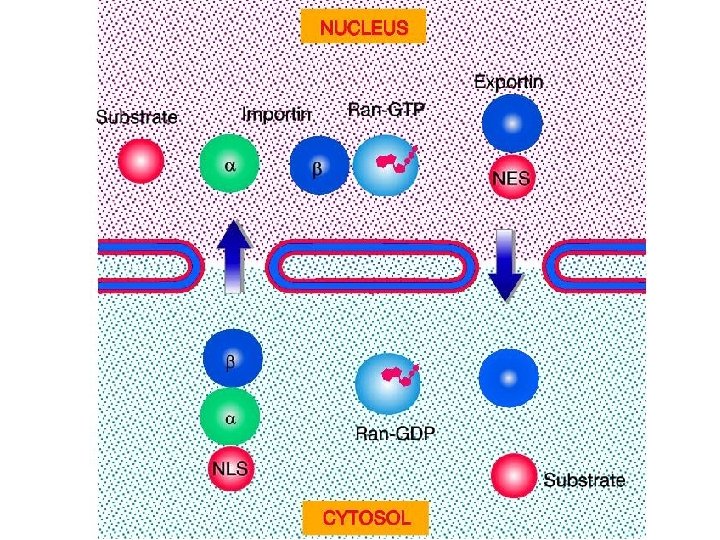
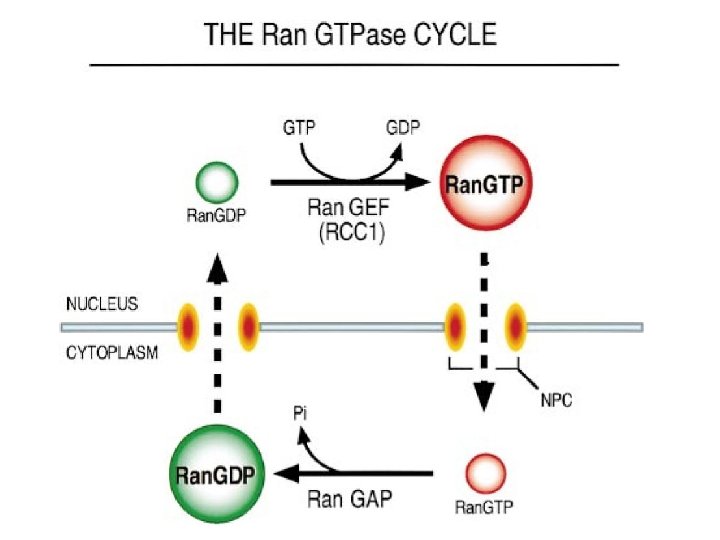
Transport through the nuclear pores • The NLS and NES consist of short sequences that are necessary and sufficient for proteins to be transported through the nuclear pores. • Transport receptors have the dual properties of recognizing NLS or NES sequences and binding to the nuclear pore. • The direction of transport is controlled by the state of the monomeric G protein, Ran. • The nucleus contains Ran-GTP, which stabilizes export complexes, while the cytosol contains Ran-GDP, which stabilizes import complexes. • The mechanism of movement does not involve a motor.

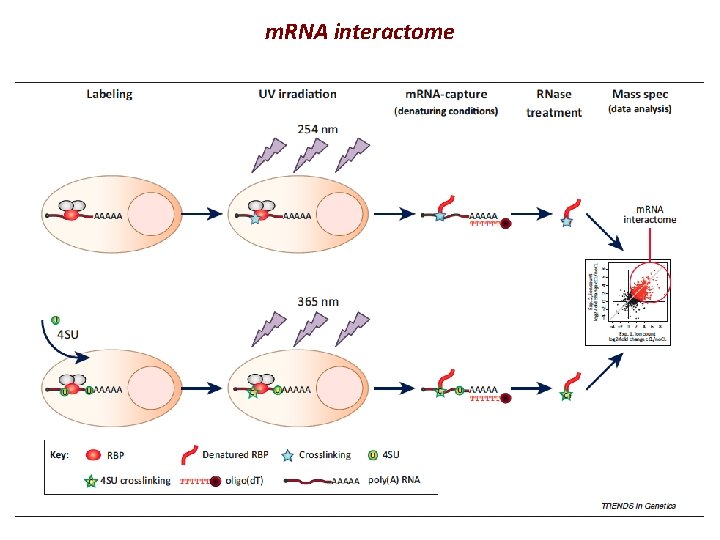
m. RNA interactome

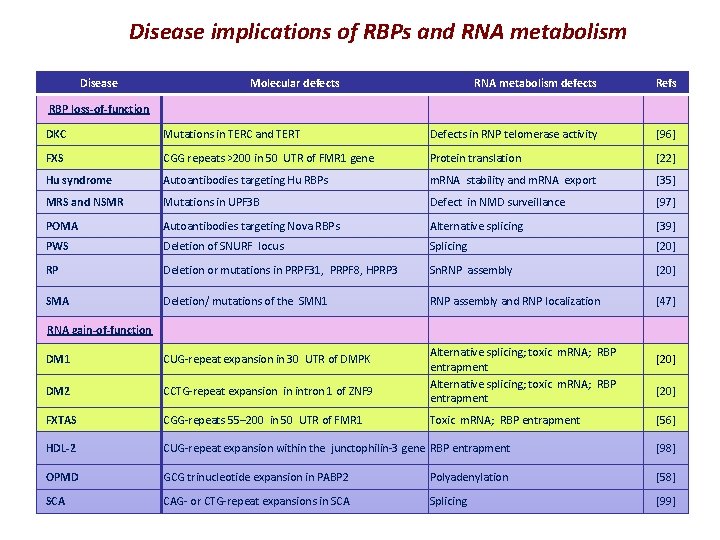
Disease implications of RBPs and RNA metabolism Disease Molecular defects RNA metabolism defects Refs RBP loss-of-function DKC Mutations in TERC and TERT Defects in RNP telomerase activity [96] FXS CGG repeats >200 in 50 UTR of FMR 1 gene Protein translation [22] Hu syndrome Autoantibodies targeting Hu RBPs m. RNA stability and m. RNA export [35] MRS and NSMR Mutations in UPF 3 B Defect in NMD surveillance [97] POMA Autoantibodies targeting Nova RBPs Alternative splicing [39] PWS Deletion of SNURF locus Splicing [20] RP Deletion or mutations in PRPF 31, PRPF 8, HPRP 3 Sn. RNP assembly [20] SMA Deletion/ mutations of the SMN 1 RNP assembly and RNP localization [47] RNA gain-of-function Alternative splicing; toxic m. RNA; RBP entrapment DM 1 CUG-repeat expansion in 30 UTR of DMPK DM 2 CCTG-repeat expansion in intron 1 of ZNF 9 FXTAS CGG-repeats 55– 200 in 50 UTR of FMR 1 HDL-2 CUG-repeat expansion within the junctophilin-3 gene RBP entrapment [98] OPMD GCG trinucleotide expansion in PABP 2 Polyadenylation [58] SCA CAG- or CTG-repeat expansions in SCA Splicing [99] Toxic m. RNA; RBP entrapment [20] [56]

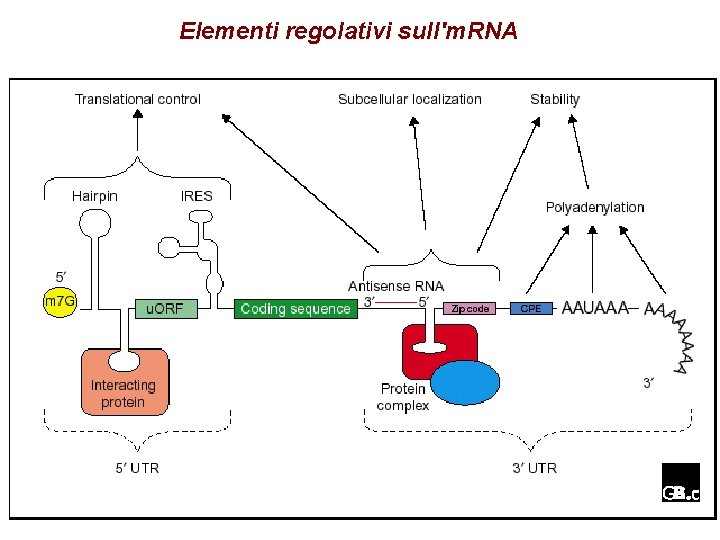
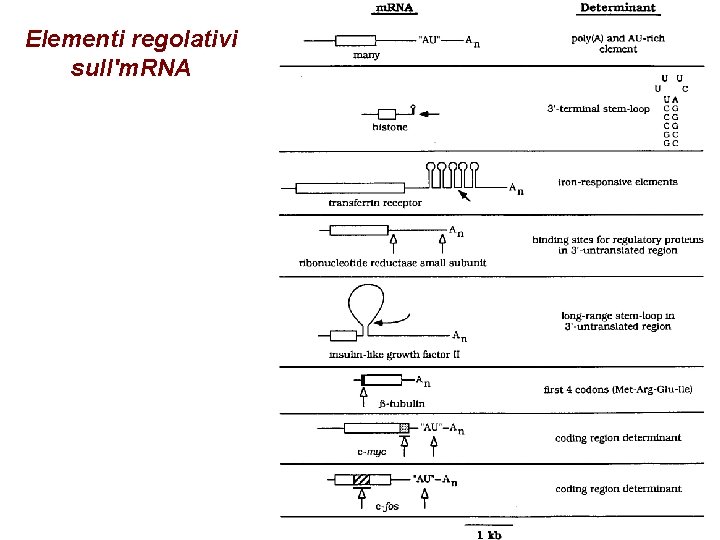
Elementi regolativi sull'm. RNA


Elementi regolativi sull'm. RNA

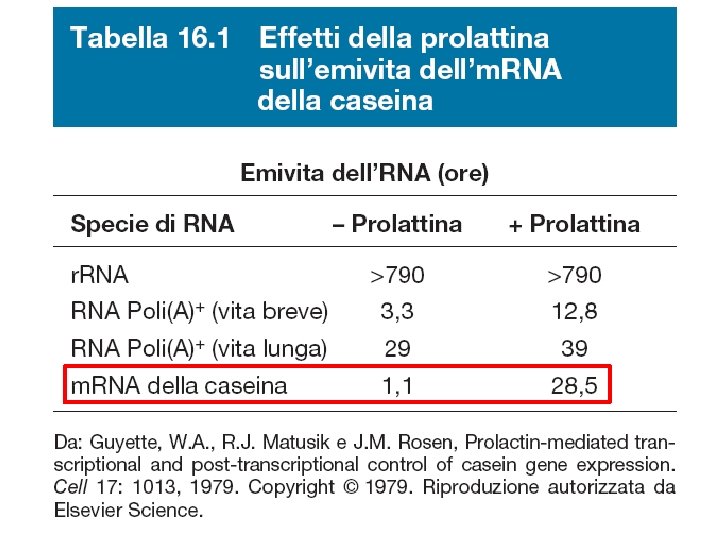
Fattori che influenzano la stabilità dell'm. RNA

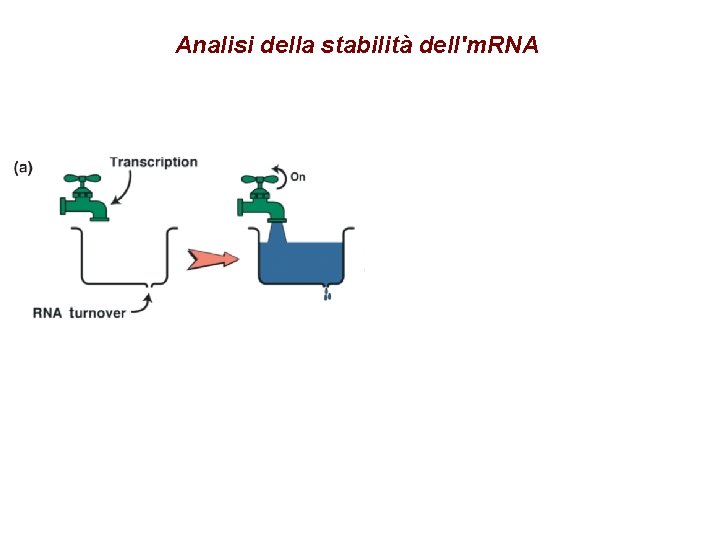
Analisi della stabilità dell'm. RNA

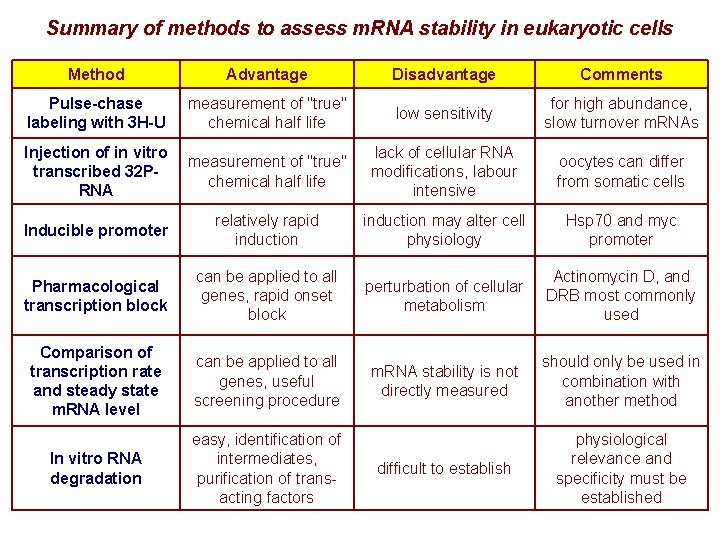
Summary of methods to assess m. RNA stability in eukaryotic cells Method Advantage Disadvantage Comments Pulse-chase labeling with 3 H-U measurement of "true" chemical half life low sensitivity for high abundance, slow turnover m. RNAs Injection of in vitro transcribed 32 PRNA measurement of "true" chemical half life lack of cellular RNA modifications, labour intensive oocytes can differ from somatic cells Inducible promoter relatively rapid induction may alter cell physiology Hsp 70 and myc promoter Pharmacological transcription block can be applied to all genes, rapid onset block perturbation of cellular metabolism Actinomycin D, and DRB most commonly used Comparison of transcription rate and steady state m. RNA level can be applied to all genes, useful screening procedure m. RNA stability is not directly measured should only be used in combination with another method In vitro RNA degradation easy, identification of intermediates, purification of transacting factors difficult to establish physiological relevance and specificity must be established

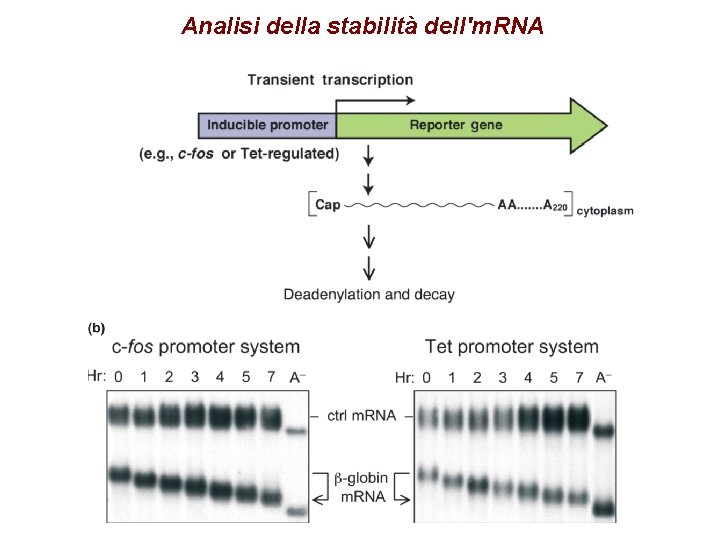
Analisi della stabilità dell'm. RNA

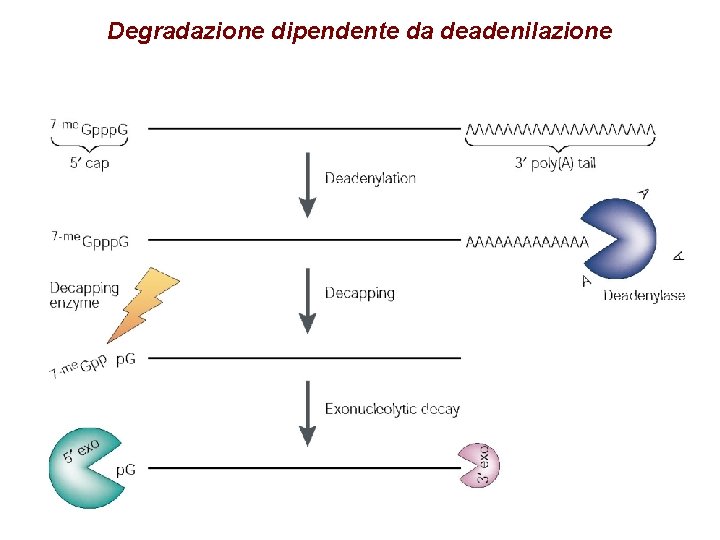
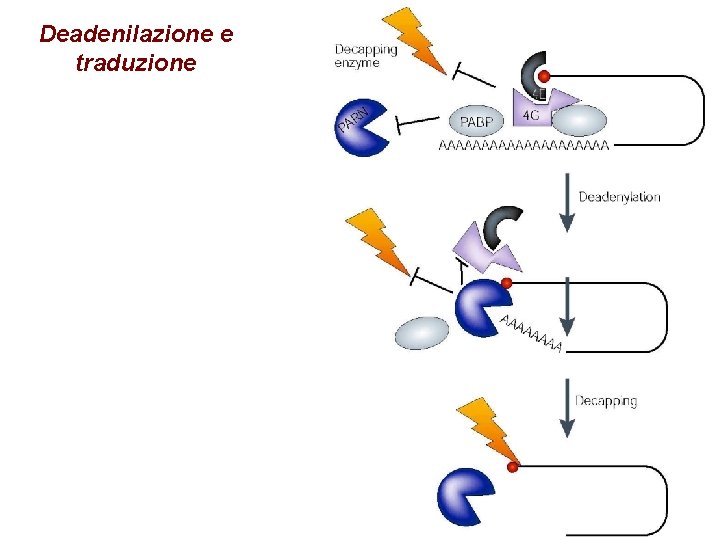
Degradazione dipendente da deadenilazione

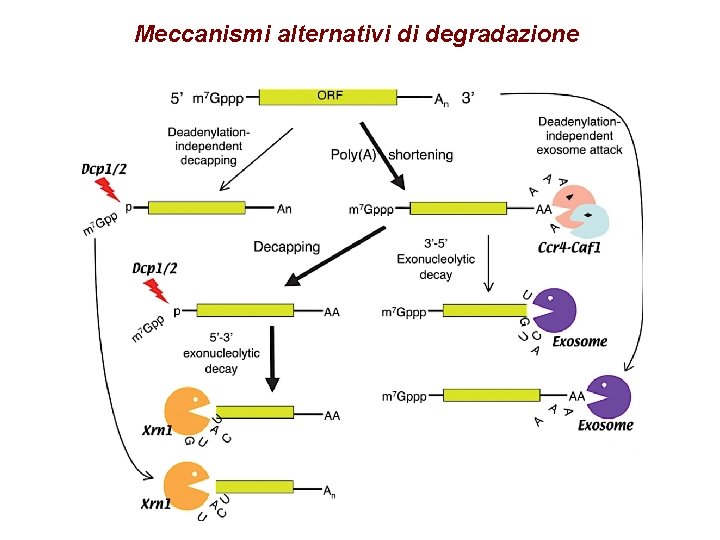
Meccanismi alternativi di degradazione

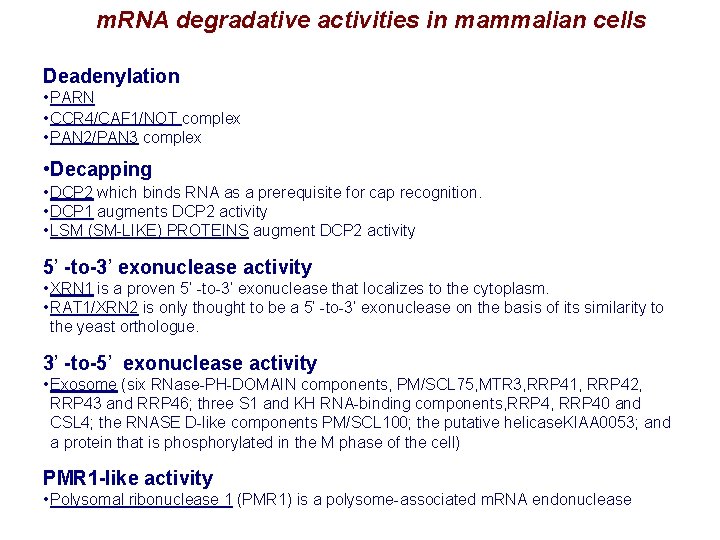
m. RNA degradative activities in mammalian cells Deadenylation • PARN • CCR 4/CAF 1/NOT complex • PAN 2/PAN 3 complex • Decapping • DCP 2 which binds RNA as a prerequisite for cap recognition. • DCP 1 augments DCP 2 activity • LSM (SM-LIKE) PROTEINS augment DCP 2 activity 5’ -to-3’ exonuclease activity • XRN 1 is a proven 5’ -to-3’ exonuclease that localizes to the cytoplasm. • RAT 1/XRN 2 is only thought to be a 5’ -to-3’ exonuclease on the basis of its similarity to the yeast orthologue. 3’ -to-5’ exonuclease activity • Exosome (six RNase-PH-DOMAIN components, PM/SCL 75, MTR 3, RRP 41, RRP 42, RRP 43 and RRP 46; three S 1 and KH RNA-binding components, RRP 40 and CSL 4; the RNASE D-like components PM/SCL 100; the putative helicase. KIAA 0053; and a protein that is phosphorylated in the M phase of the cell) PMR 1 -like activity • Polysomal ribonuclease 1 (PMR 1) is a polysome-associated m. RNA endonuclease

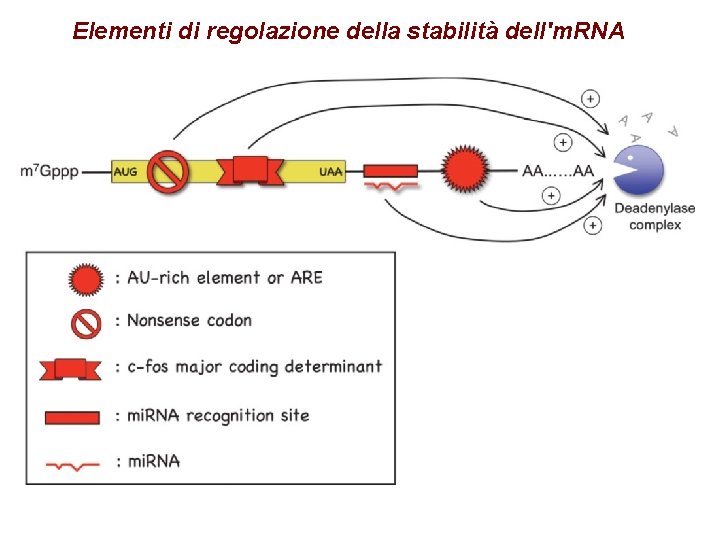
Elementi di regolazione della stabilità dell'm. RNA

Deadenilazione e traduzione

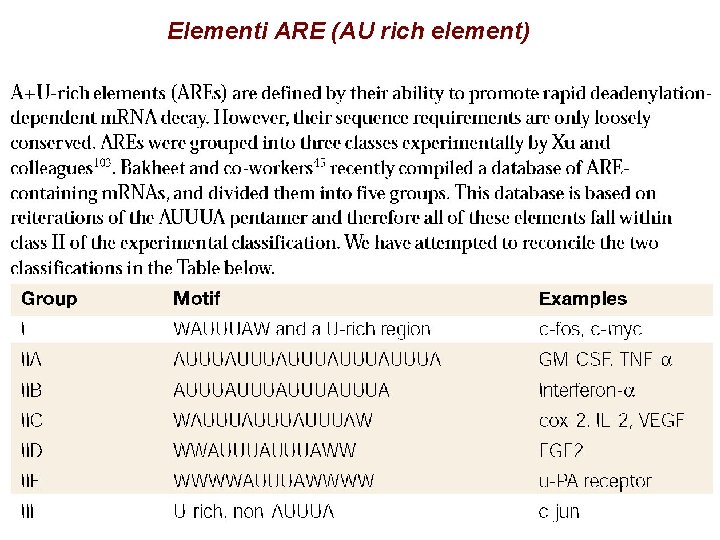
Elementi ARE (AU rich element)

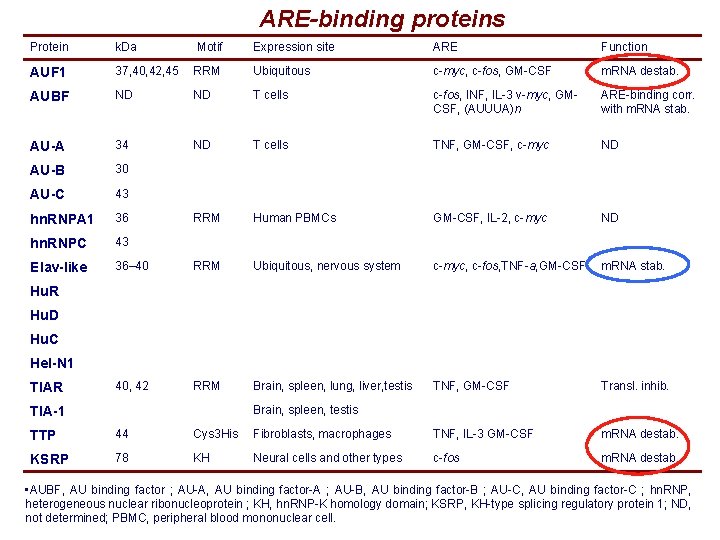
ARE-binding proteins Protein k. Da Motif Expression site ARE Function AUF 1 37, 40, 42, 45 RRM Ubiquitous c-myc, c-fos, GM-CSF m. RNA destab. AUBF ND ND T cells c-fos, INF, IL-3 v-myc, GMCSF, (AUUUA)n ARE-binding corr. with m. RNA stab. AU-A 34 ND T cells TNF, GM-CSF, c-myc ND AU-B 30 AU-C 43 hn. RNPA 1 36 RRM Human PBMCs GM-CSF, IL-2, c-myc ND hn. RNPC 43 Elav-like 36– 40 RRM Ubiquitous, nervous system c-myc, c-fos, TNF-a, GM-CSF m. RNA stab. 40, 42 RRM Brain, spleen, lung, liver, testis TNF, GM-CSF Transl. inhib. Hu. R Hu. D Hu. C Hel-N 1 TIAR Brain, spleen, testis TIA-1 TTP 44 Cys 3 His Fibroblasts, macrophages TNF, IL-3 GM-CSF m. RNA destab. KSRP 78 KH Neural cells and other types c-fos m. RNA destab. • AUBF, AU binding factor ; AU-A, AU binding factor-A ; AU-B, AU binding factor-B ; AU-C, AU binding factor-C ; hn. RNP, heterogeneous nuclear ribonucleoprotein ; KH, hn. RNP-K homology domain; KSRP, KH-type splicing regulatory protein 1; ND, not determined; PBMC, peripheral blood mononuclear cell.

ARE-binding proteins

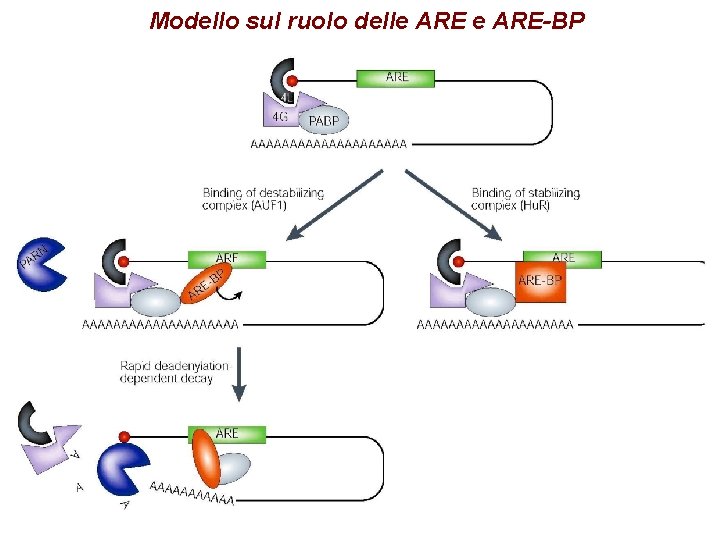
Modello sul ruolo delle ARE-BP

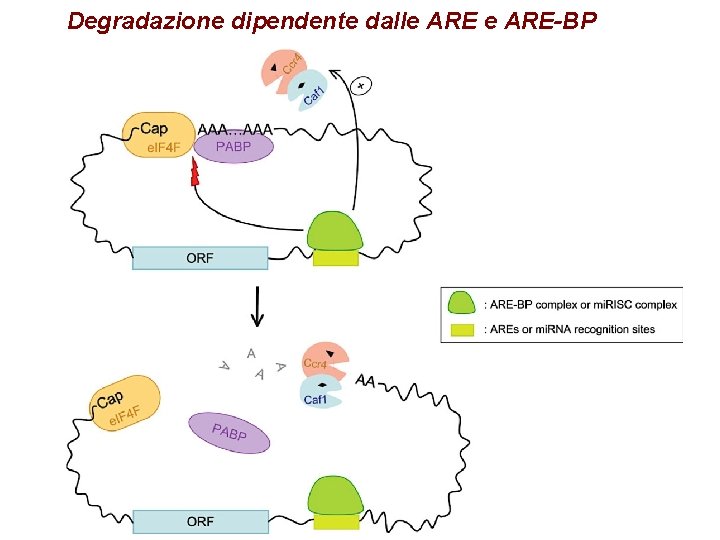
Degradazione dipendente dalle ARE-BP

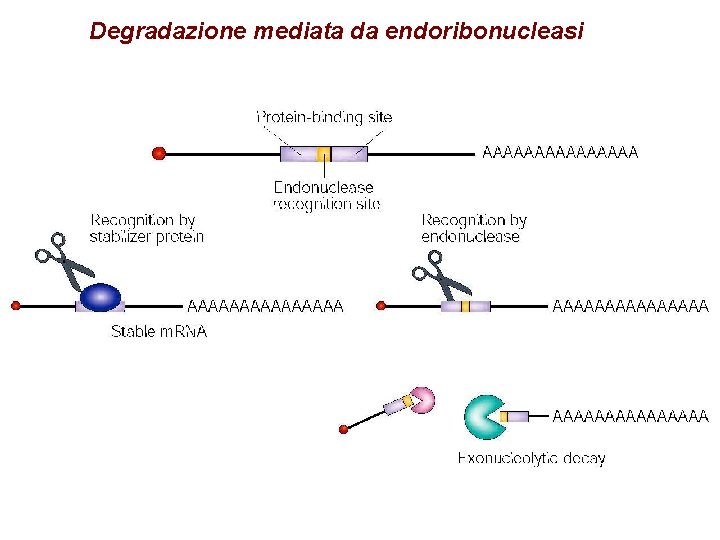
Degradazione mediata da endoribonucleasi
 Xna visual studio 2017
Xna visual studio 2017 Xbox xna
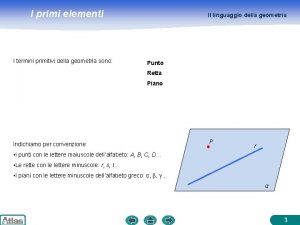
Xbox xna I termini primitivi sono elementi
I termini primitivi sono elementi Trasporto attivo diretto e indiretto
Trasporto attivo diretto e indiretto Trasporto attivo indiretto
Trasporto attivo indiretto Somme algebriche di radicali
Somme algebriche di radicali Mobilizzazione atraumatica definizione
Mobilizzazione atraumatica definizione Foglio rosa trasporto cavalli
Foglio rosa trasporto cavalli Trasporto isoidrico della co2
Trasporto isoidrico della co2 Catena di trasporto degli elettroni zanichelli
Catena di trasporto degli elettroni zanichelli Trasporto di cristo
Trasporto di cristo Mezzi di trasporto via aria
Mezzi di trasporto via aria Cellula procariote
Cellula procariote Membrana plasmatica
Membrana plasmatica Proteine di trasporto
Proteine di trasporto Allegato xlvi del d.lgs. 81/08 coronavirus
Allegato xlvi del d.lgs. 81/08 coronavirus Caratteristiche del linguaggio umano
Caratteristiche del linguaggio umano Site:slidetodoc.com
Site:slidetodoc.com Transfer rna
Transfer rna Rna protein synthesis
Rna protein synthesis Blank a spectrophotometer
Blank a spectrophotometer Messenger rna sequence
Messenger rna sequence Sam file
Sam file Rna polymerase 1 2 3
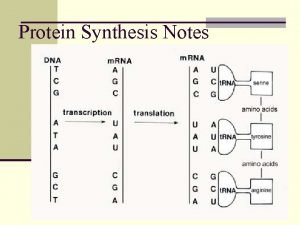
Rna polymerase 1 2 3 Dna mrna trna amino acid chart
Dna mrna trna amino acid chart Rna catalyst
Rna catalyst Section 12 3 rna and protein synthesis
Section 12 3 rna and protein synthesis Protein synthesis
Protein synthesis Rasi rna
Rasi rna Rna wheel
Rna wheel Rna translation
Rna translation