Protein Coexpression Network Analysis in Alzheimers Disease Nick


- Slides: 24

Protein Co-expression Network Analysis in Alzheimer’s Disease Nick Seyfried Department of Biochemistry Alzheimer’s Disease Research Center Director, Emory Integrated Proteomics Core Emory University, School of Medicine

Proteomics in Alzheimer’s Disease (AD) Specific Goal • Develop accurate and precise method to quantify proteins in human postmortem brain tissue Ultimate goal: goal Can we use proteomics to better define preclinical stages of AD and key molecular pathways that associate with cognitive decline?

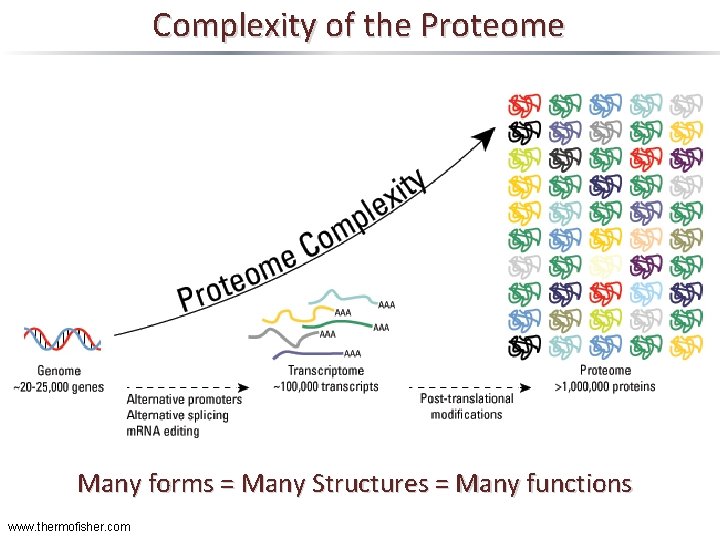
Complexity of the Proteome Many forms = Many Structures = Many functions www. thermofisher. com

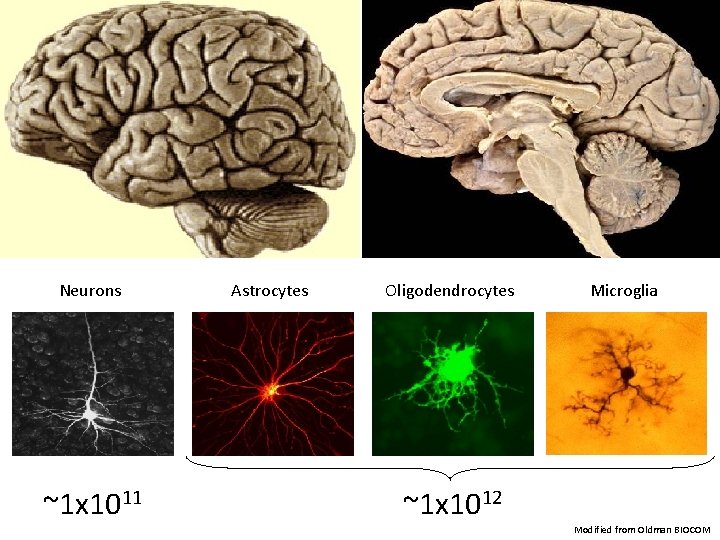
Neurons ~1 x 1011 Astrocytes Oligodendrocytes Microglia ~1 x 1012 Modified from Oldman BIOCOM

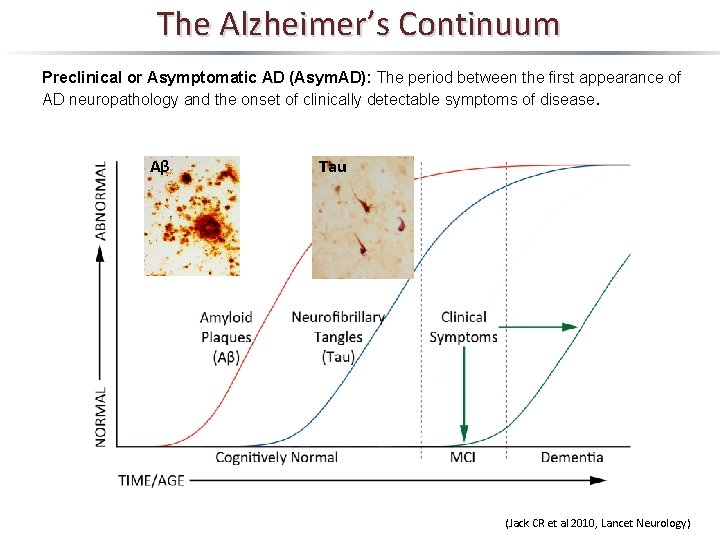
The Alzheimer’s Continuum Preclinical or Asymptomatic AD (Asym. AD): The period between the first appearance of AD neuropathology and the onset of clinically detectable symptoms of disease. Aβ Tau (Jack CR et al 2010, Lancet Neurology)

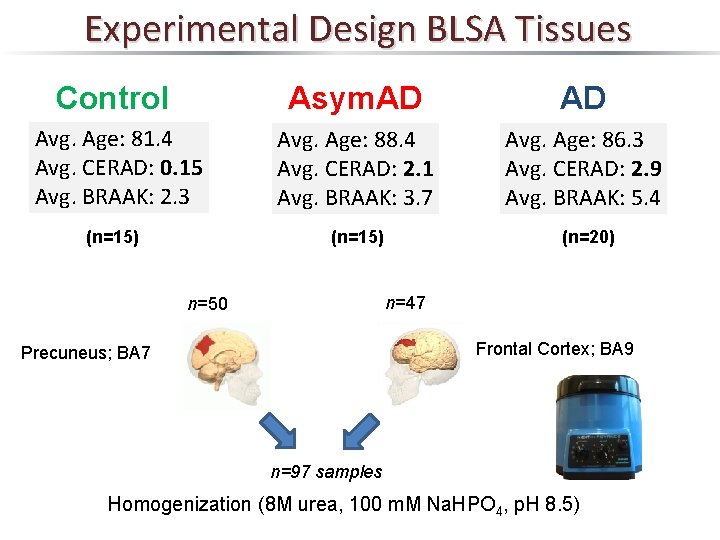
Experimental Design BLSA Tissues Control Asym. AD AD Avg. Age: 81. 4 Avg. CERAD: 0. 15 Avg. BRAAK: 2. 3 Avg. Age: 88. 4 Avg. CERAD: 2. 1 Avg. BRAAK: 3. 7 Avg. Age: 86. 3 Avg. CERAD: 2. 9 Avg. BRAAK: 5. 4 (n=15) (n=20) (n=15) n=47 n=50 Frontal Cortex; BA 9 Precuneus; BA 7 n=97 samples Homogenization (8 M urea, 100 m. M Na. HPO 4, p. H 8. 5)

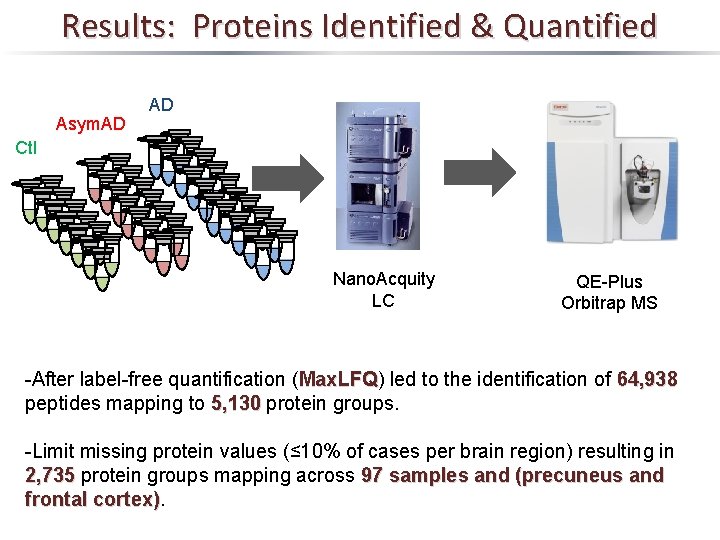
Results: Proteins Identified & Quantified Asym. AD AD Ctl Nano. Acquity LC QE-Plus Orbitrap MS -After label-free quantification (Max. LFQ) Max. LFQ led to the identification of 64, 938 peptides mapping to 5, 130 protein groups. -Limit missing protein values (≤ 10% of cases per brain region) resulting in 2, 735 protein groups mapping across 97 samples and (precuneus and frontal cortex)

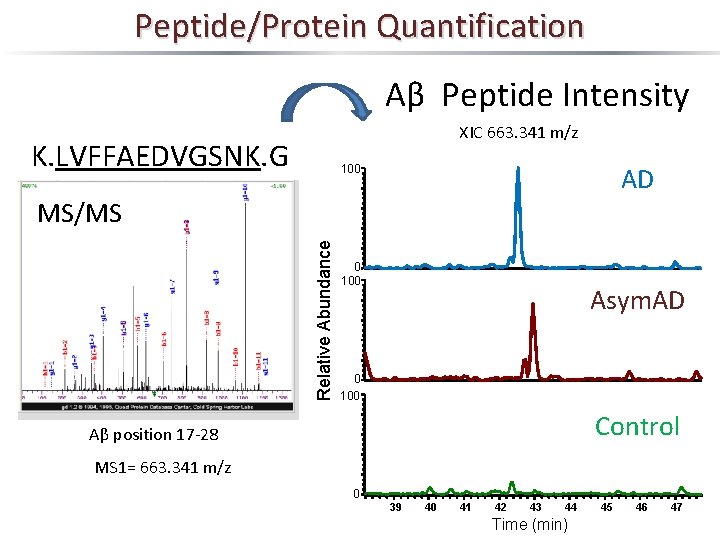
Peptide/Protein Quantification Aβ Peptide Intensity XIC 663. 341 m/z K. LVFFAEDVGSNK. G AD 100 Relative Abundance MS/MS 0 100 Asym. AD 0 100 Control Aβ position 17 -28 MS 1= 663. 341 m/z 0 39 40 41 42 43 44 Time (min) 45 46 47

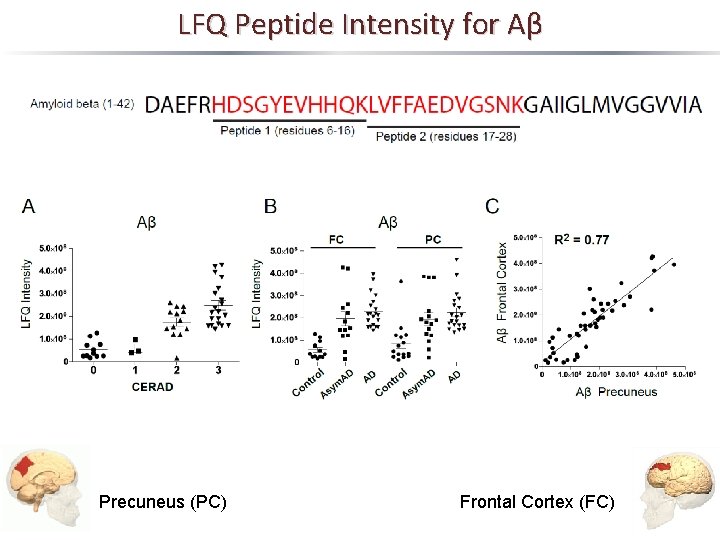
LFQ Peptide Intensity for Aβ Precuneus (PC) Frontal Cortex (FC)

Wei. Ghted Co-expression Network Analysis (WGCNA) • Understand the “system” instead of reporting a list of individual parts • Focus on modules or groups of proteins as opposed to individual proteins • Module identification is an essential step towards understanding the whole network architecture Assess protein expression patterns across samples Define modules or networks of highly correlated proteins Define biological function of these networks and key “hub” proteins Assess module-trait Relationships

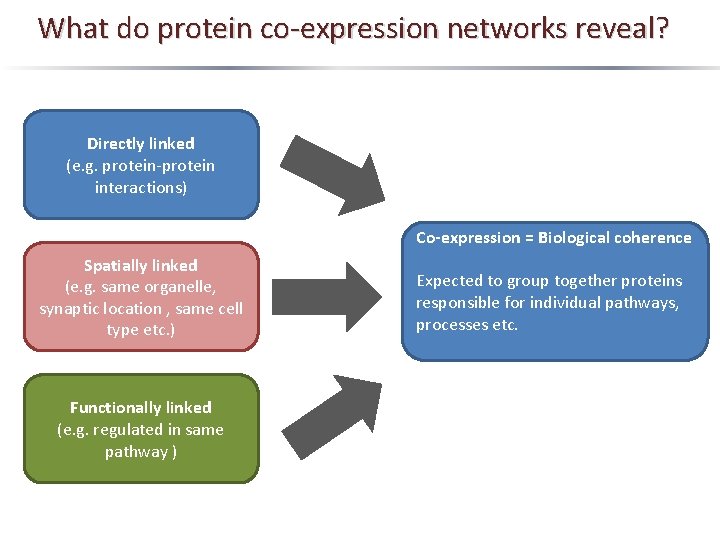
What do protein co-expression networks reveal? Directly linked (e. g. protein-protein interactions) Co-expression = Biological coherence Spatially linked (e. g. same organelle, synaptic location , same cell type etc. ) Functionally linked (e. g. regulated in same pathway ) Expected to group together proteins responsible for individual pathways, processes etc.

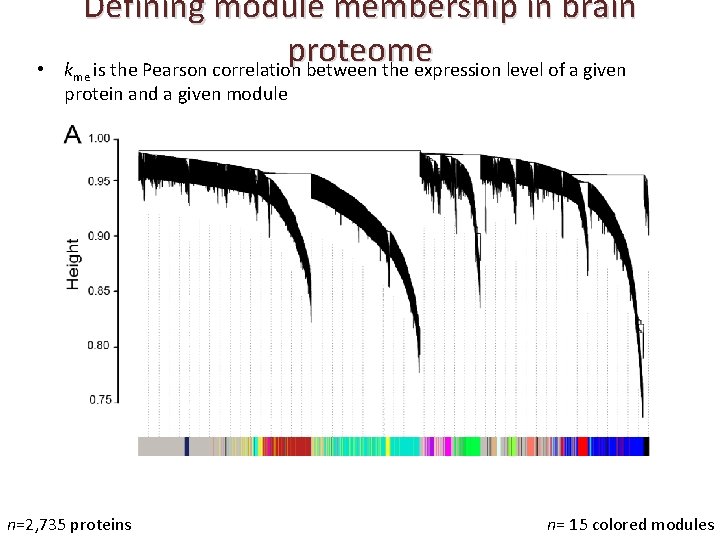
• Defining module membership in brain proteome k is the Pearson correlation between the expression level of a given me protein and a given module n=2, 735 proteins n= 15 colored modules

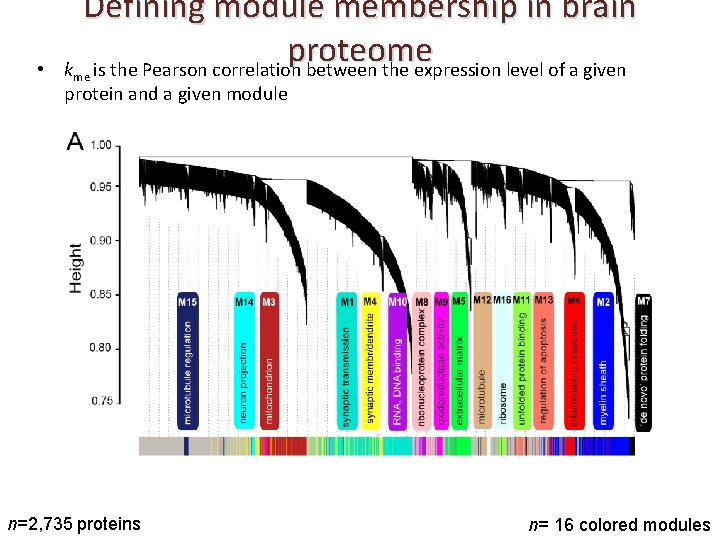
• Defining module membership in brain proteome k is the Pearson correlation between the expression level of a given me protein and a given module n=2, 735 proteins n= 16 colored modules

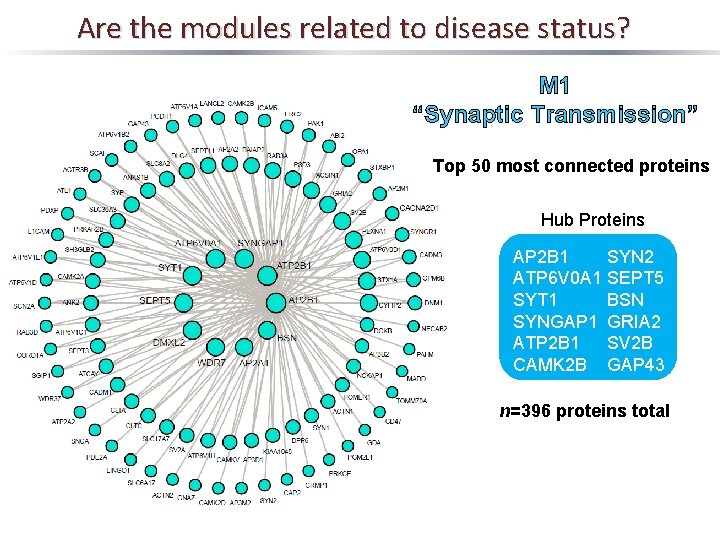
Are the modules related to disease status? M 1 “Synaptic Transmission” Top 50 most connected proteins Hub Proteins AP 2 B 1 ATP 6 V 0 A 1 SYT 1 SYNGAP 1 ATP 2 B 1 CAMK 2 B SYN 2 SEPT 5 BSN GRIA 2 SV 2 B GAP 43 n=396 proteins total

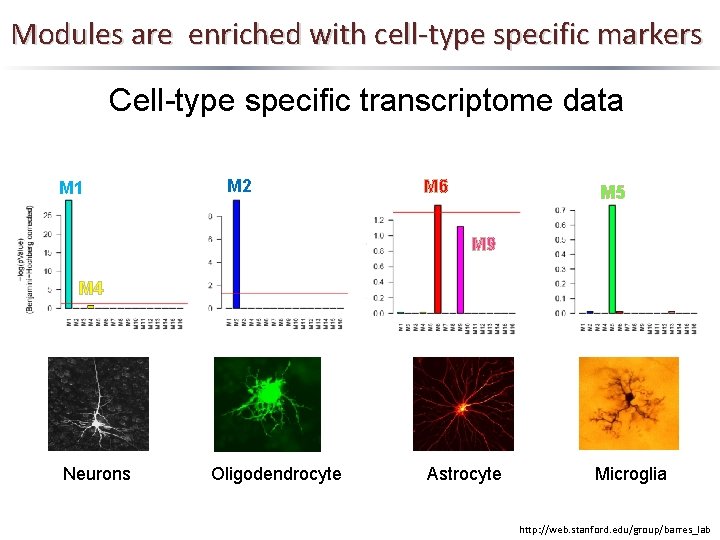
Modules are enriched with cell-type specific markers Cell-type specific transcriptome data M 1 M 2 M 6 M 5 M 9 M 4 Neurons Oligodendrocyte Astrocyte Microglia http: //web. stanford. edu/group/barres_lab

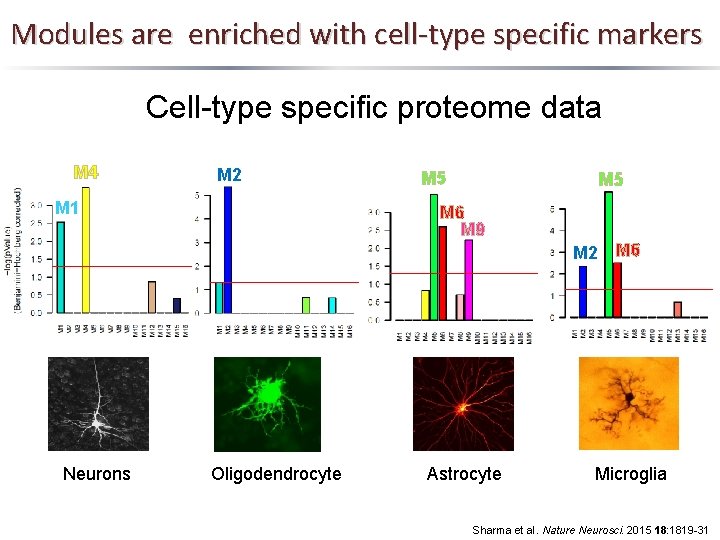
Modules are enriched with cell-type specific markers Cell-type specific proteome data M 4 M 2 M 1 M 5 M 6 M 9 M 2 M 6 Neurons Oligodendrocyte Astrocyte Microglia Sharma et al. Nature Neurosci. 2015 18: 1819 -31

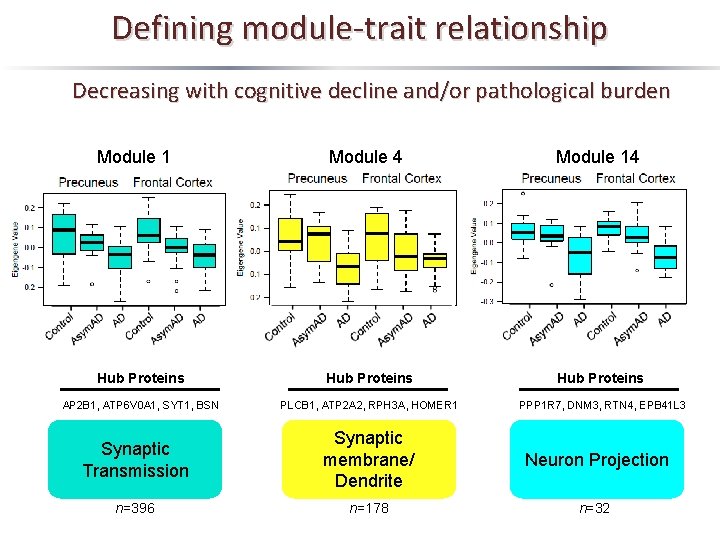
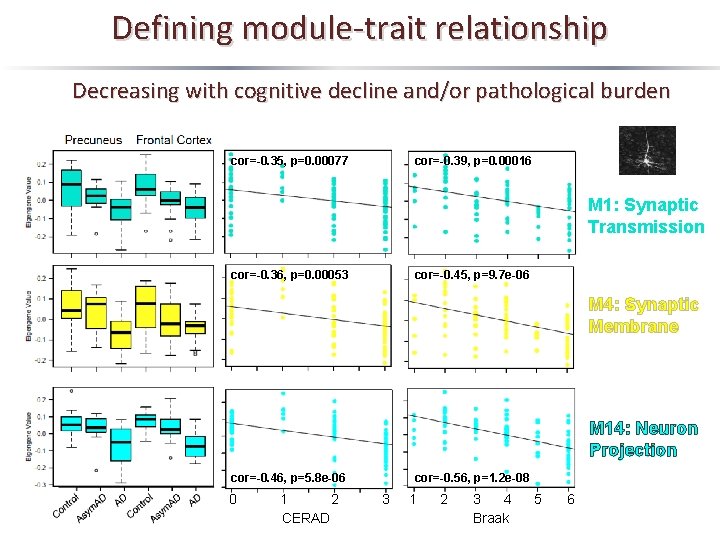
Defining module-trait relationship Decreasing with cognitive decline and/or pathological burden Module 1 Module 4 Module 14 Hub Proteins AP 2 B 1, ATP 6 V 0 A 1, SYT 1, BSN PLCB 1, ATP 2 A 2, RPH 3 A, HOMER 1 PPP 1 R 7, DNM 3, RTN 4, EPB 41 L 3 Synaptic Transmission Synaptic membrane/ Dendrite Neuron Projection n=396 n=178 n=32

Defining module-trait relationship Decreasing with cognitive decline and/or pathological burden cor=-0. 35, p=0. 00077 cor=-0. 39, p=0. 00016 M 1: Synaptic Transmission cor=-0. 36, p=0. 00053 cor=-0. 45, p=9. 7 e-06 M 4: Synaptic Membrane M 14: Neuron Projection cor=-0. 46, p=5. 8 e-06 0 1 2 CERAD cor=-0. 56, p=1. 2 e-08 3 1 2 3 4 Braak 5 6

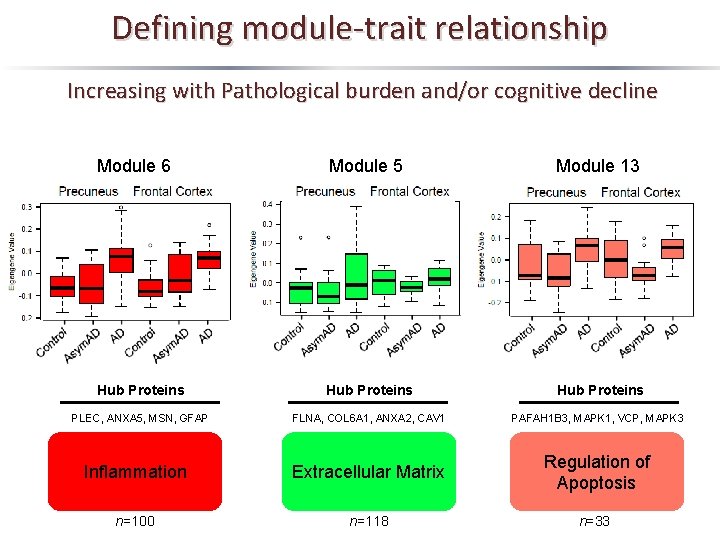
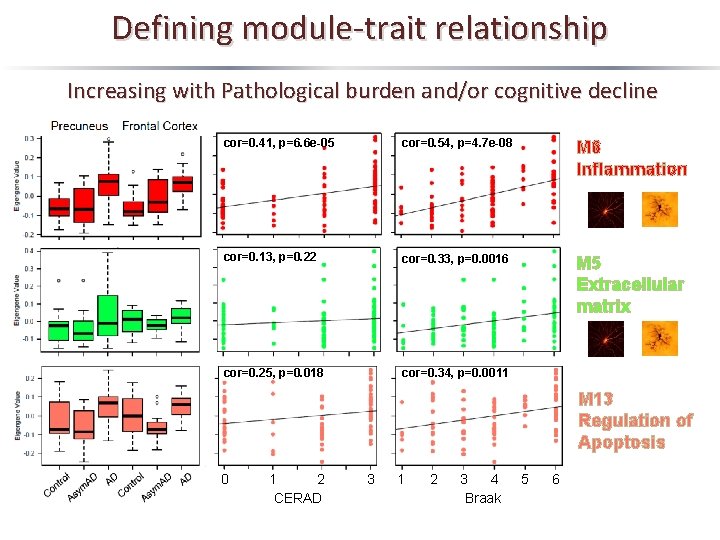
Defining module-trait relationship Increasing with Pathological burden and/or cognitive decline Module 6 Module 5 Module 13 Hub Proteins PLEC, ANXA 5, MSN, GFAP FLNA, COL 6 A 1, ANXA 2, CAV 1 PAFAH 1 B 3, MAPK 1, VCP, MAPK 3 Inflammation Extracellular Matrix Regulation of Apoptosis n=100 n=118 n=33

Defining module-trait relationship Increasing with Pathological burden and/or cognitive decline cor=0. 41, p=6. 6 e-05 cor=0. 54, p=4. 7 e-08 M 6 Inflammation cor=0. 13, p=0. 22 cor=0. 33, p=0. 0016 M 5 Extracellular matrix cor=0. 25, p=0. 018 cor=0. 34, p=0. 0011 M 13 Regulation of Apoptosis 0 1 2 CERAD 3 1 2 3 4 Braak 5 6

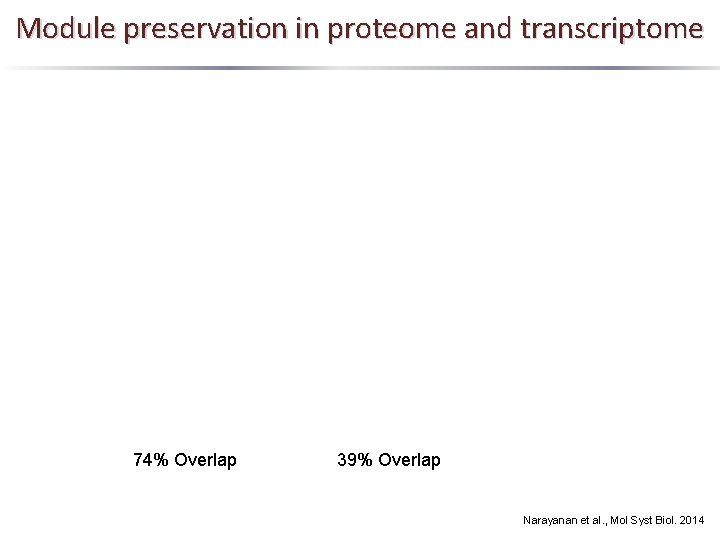
Module preservation in proteome and transcriptome 74% Overlap 39% Overlap Narayanan et al. , Mol Syst Biol. 2014

Convergent modules related to Cell-types display strong RNA-protein Correlation

Conclusions and Implications • Modules represent specific cell types and functional complexes with links to cellular processes • Protein co-expression analysis defines biological networks correlated to neuropathology and cognitive decline in AD • Future Directions- ROS/MAP proteome and transcriptome integration • Targeted brain networks in CSF as a path towards biomarkers Systems level analysis of the proteome has potential to reveal cell-type driven networks and therapeutic targets in AD

Acknowledgements Allan Levey Jim Lah Eric Dammer Ian Diner Duc Duong Tram Nguyen Ranjita Betarbet Measho Abreha Juan C. Troncoso Giovanni Coppola Dan Geschwind Vivek Swarup Marla Gearing Qiudong Deng Chad Hales Divya Nandakumar Lingyan Ping Luming Yin Isaac Bishof Madhav Thambisetty Josh Shulman Funding: