Phylogenomics JerMing Hu Plant Systematics and Evolution Group











- Slides: 70

Phylogenomics Jer-Ming Hu 胡哲明 Plant Systematics and Evolution Group, NTU 台大植物系統分類與演化研究室 本著作除另有註明外,採取創用CC「姓名標示- 非商業性-相同方式分享」台灣 3. 0版授權釋出 Unless noted, the course materials are licensed under Creative Commons Attribution-Non. Commercial-Share. Alike 3. 0 Taiwan (CC BY-NC-SA 3. 0) 1

Phylogenomics n The uses of phylogenetic principles to make sense of genomic data n Recent reviews n Eisen, J. A. 1998. Gen. Res. 8: 163 -167 n Delsuc, F. et al. 2005. Nat. Rev. Genet. 6: 361 -375 n Koonin, E. V. 2005. Annu. Rev. Genet. 39: 309 -338 n Philippe, H. et al. 2005. Annu. Rev. Ecol. Evol. Syst. 36: 541 -562 n Boussau & Daubin (2010) TREE 25: 224 -232 2

Current approaches � Methods based on primary sequences � � Supermatrix Supertree Large-scale screening (e. g. ngs data) Methods based on whole-genome features � � � Gene content Gene order DNA strings (distribution of oligonucleotides in genomes) 3

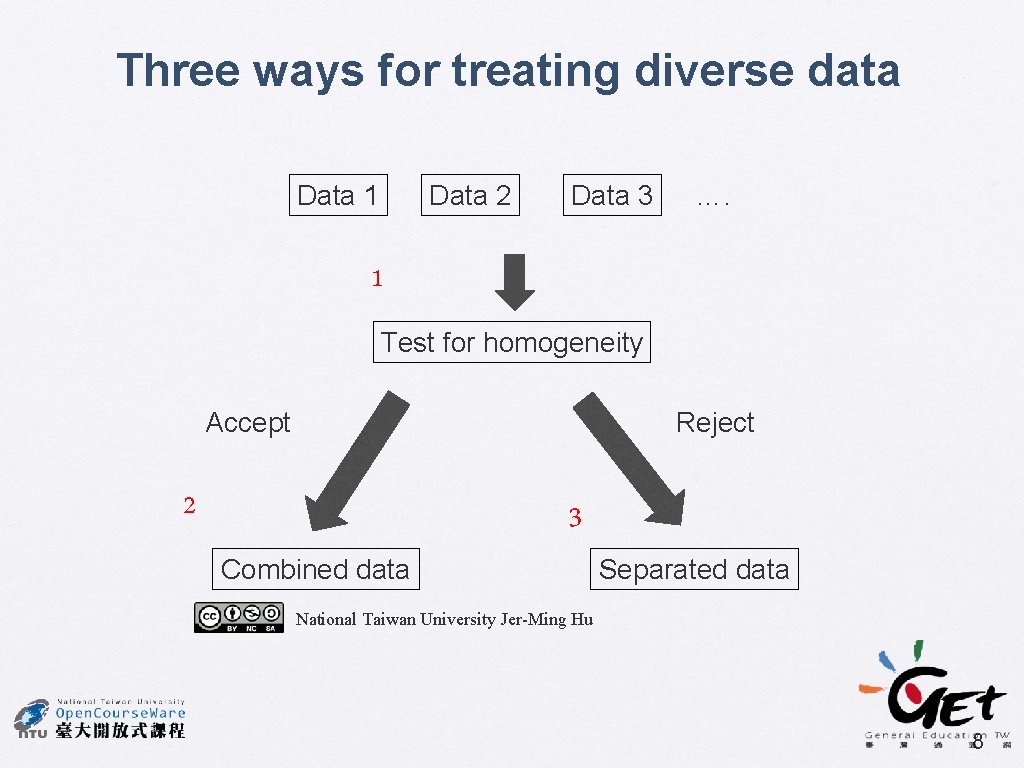
Combining data in phylogenetic analysis � Data partition � � � Different genes Coding vs. non-coding regions Among codon positions Molecular vs. morphological data Three ways to deal with partitioned data � � � Always combine (total evidence) Never combine (separate analysis) Conditional combine (perform a test to decide whether or not to combine them) Huelsenbeck, J. P. et al. (1996) TREE 11: 152 4

Separate analysis � � � Miyamoto and Fitch (1995) Tree should be estimated from each partition Congruence among different data partitions provides stronger evidence of accurate phylogeny Miyamoto, M. M. and W. M. Fitch (1995) Syst. Biol. 44: 64 -76 5

Logic of total evidence � � All independent characters should be included without a priori consideration of data partition Not only character data, but also taxa (e. g. fossil and living) should be included in a phylogenetic analysis. 6

Conditional data combination � � Bull et al. (1993), de Queiroz (1993) The partitions should be subjected to a homogeneity test before any further process. � � e. g. KH test, partition homogeneity test, likelihood heterogeneity test, etc. Potential problem with false positives � The failure to combine data sets that should have been combined Bull, J. J. et al. (1993) Syst. Biol. 42: 384 -397 de Queiroz, A. (1993) Syst. Biol. 42: 368 -372 7

Three ways for treating diverse data Data 1 Data 2 Data 3 …. 1 Test for homogeneity Accept Reject 2 3 Combined data Separated data National Taiwan University Jer-Ming Hu 8

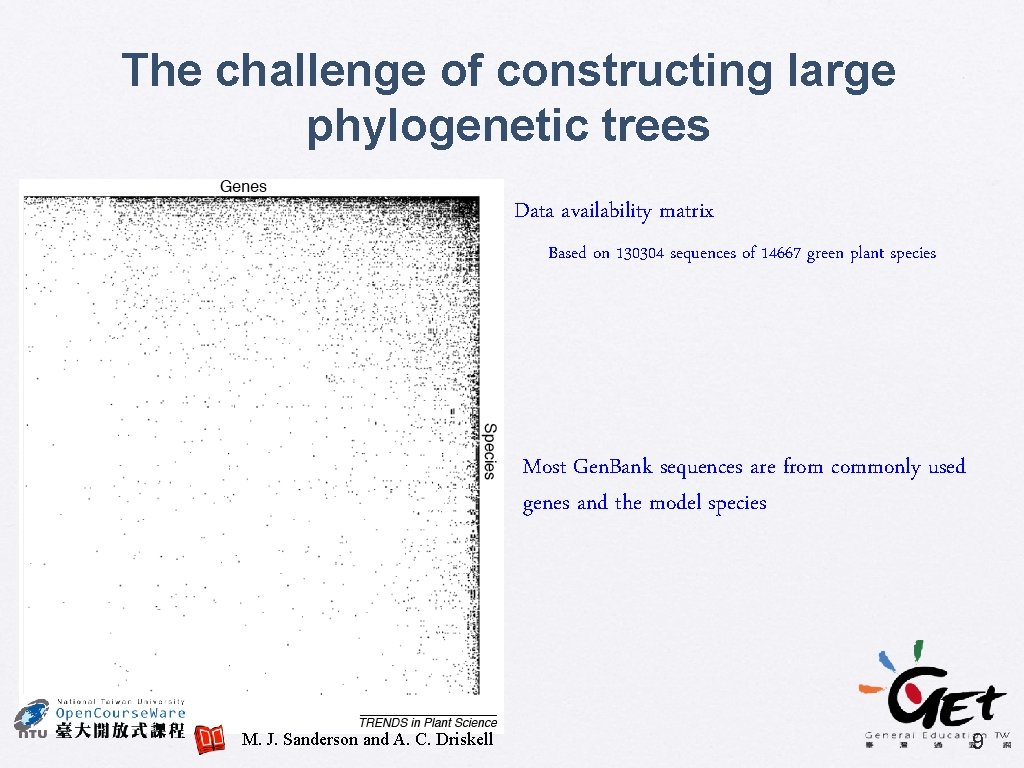
The challenge of constructing large phylogenetic trees Data availability matrix Based on 130304 sequences of 14667 green plant species Most Gen. Bank sequences are from commonly used genes and the model species M. J. Sanderson and A. C. Driskell 9

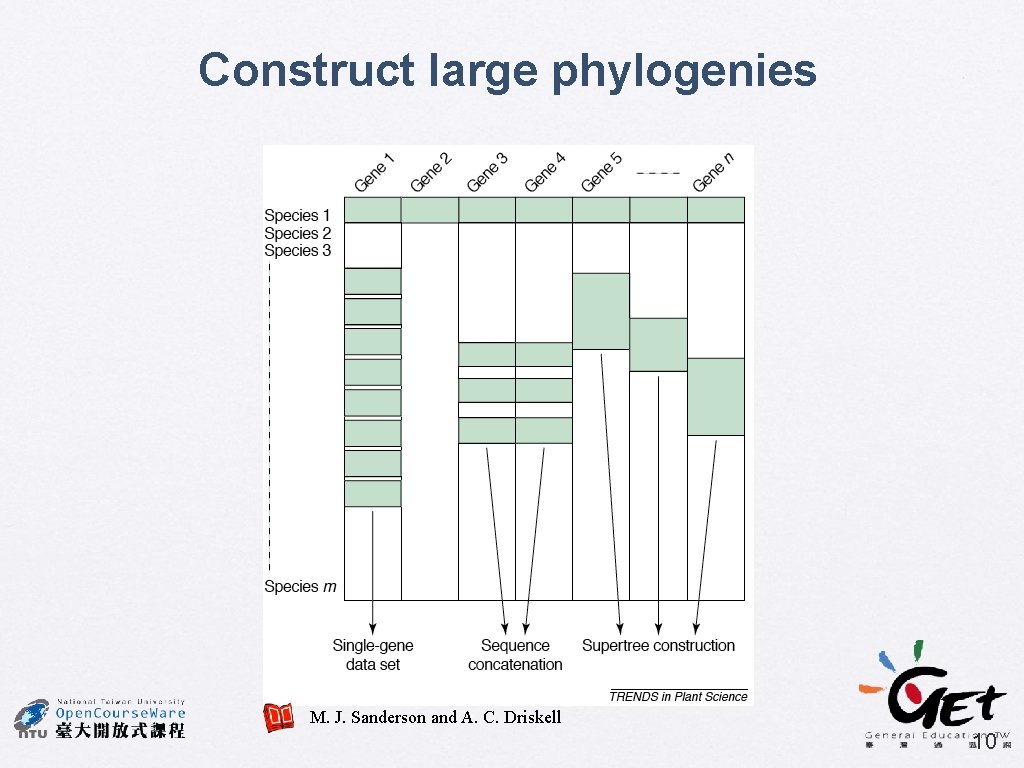
Construct large phylogenies M. J. Sanderson and A. C. Driskell 10

Problems of data concatenation � � Including multiple genes may mix phylogenetic signals from different evolutionary histories Some sequences are usually unavailable for some species and have to code them as missing data � e. g. photosynthetic genes of non-photosynthetic plants 11

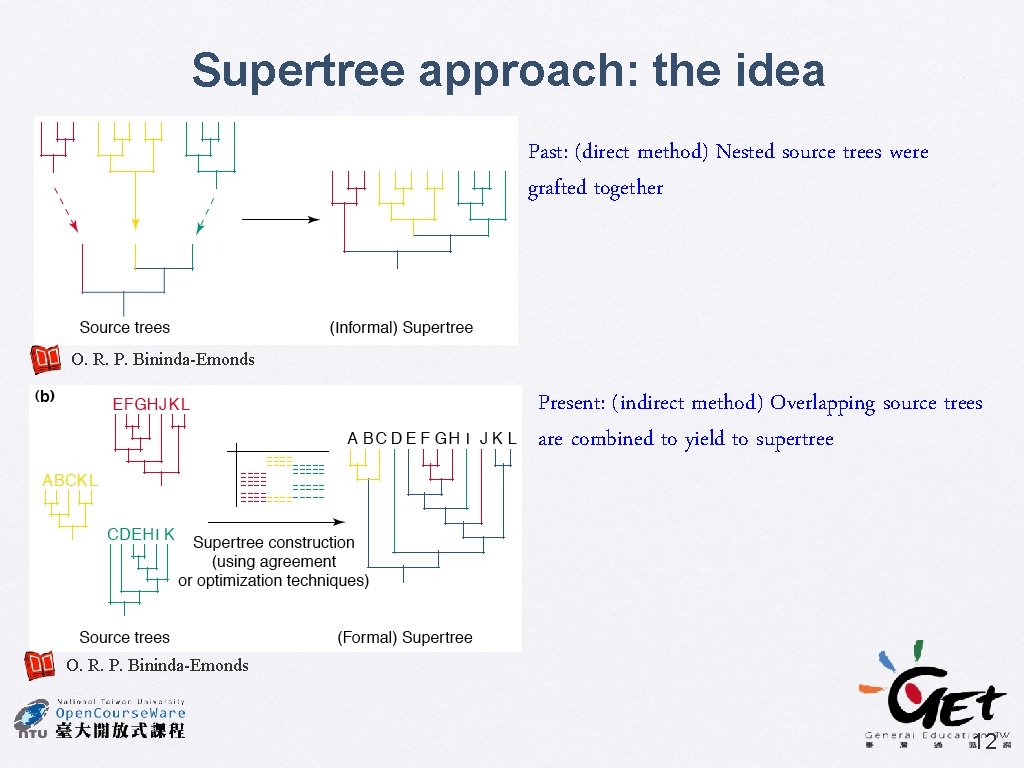
Supertree approach: the idea Past: (direct method) Nested source trees were grafted together O. R. P. Bininda-Emonds Present: (indirect method) Overlapping source trees are combined to yield to supertree O. R. P. Bininda-Emonds 12

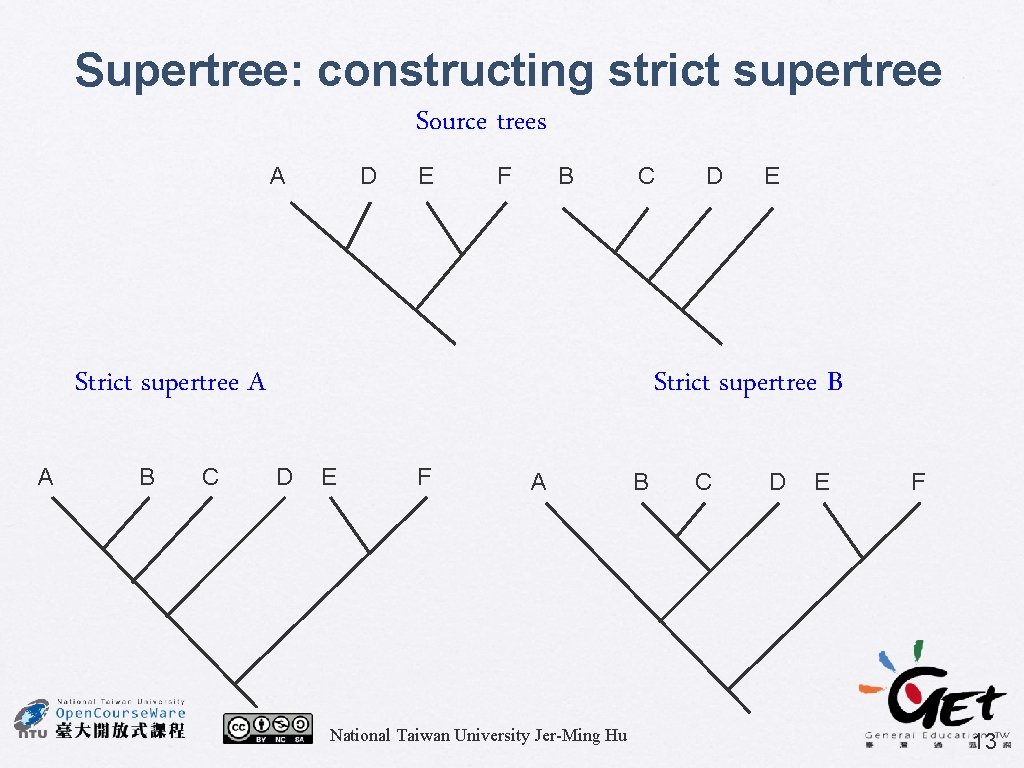
Supertree: constructing strict supertree Source trees A D E F B C Strict supertree A A B C D E Strict supertree B D E F A National Taiwan University Jer-Ming Hu B C D E F 13

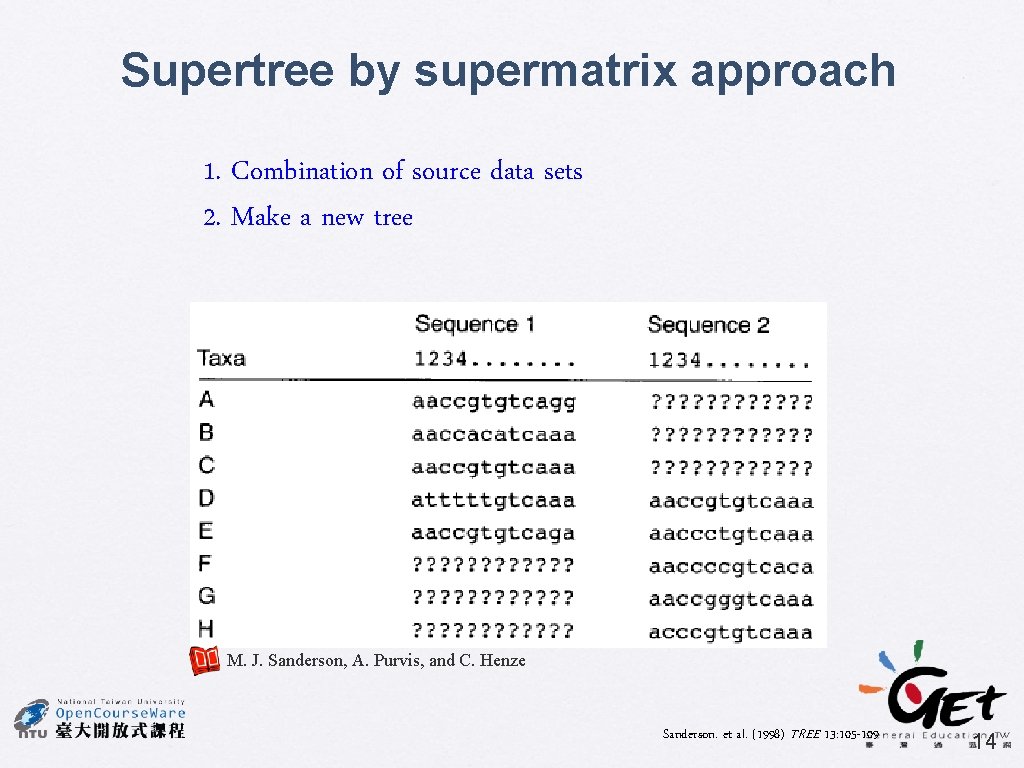
Supertree by supermatrix approach 1. Combination of source data sets 2. Make a new tree M. J. Sanderson, A. Purvis, and C. Henze Sanderson. et al. (1998) TREE 13: 105 -109 14

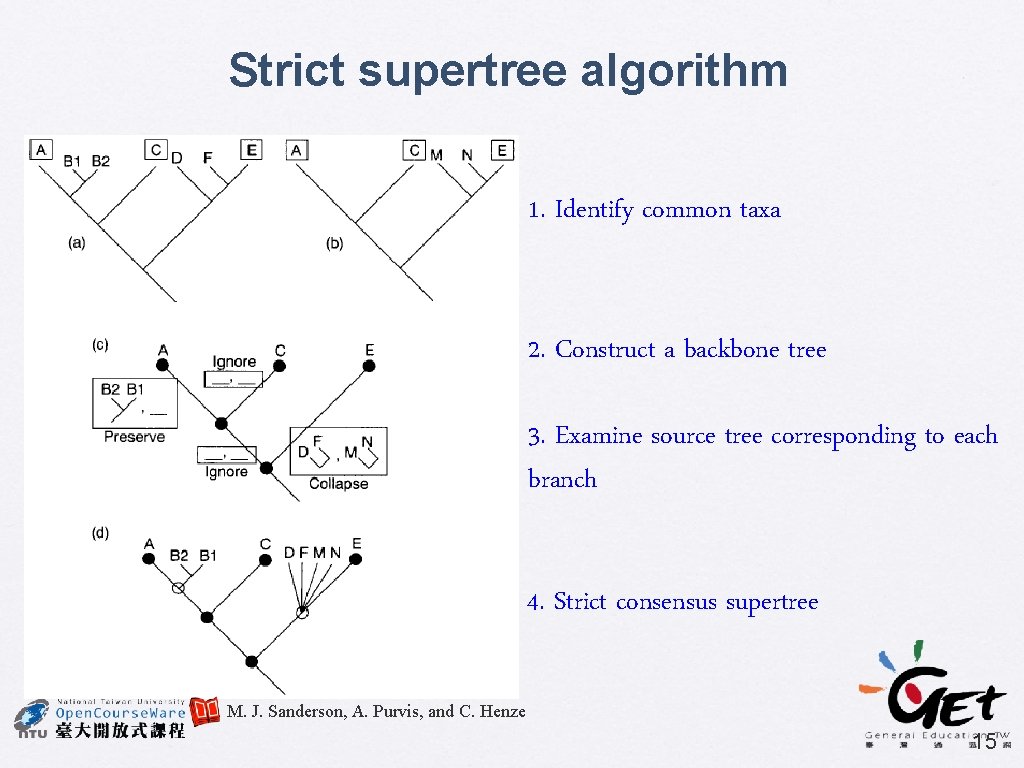
Strict supertree algorithm 1. Identify common taxa 2. Construct a backbone tree 3. Examine source tree corresponding to each branch 4. Strict consensus supertree M. J. Sanderson, A. Purvis, and C. Henze 15

Supertree by MRP � � Matrix Representation with Parsimony (MRP) Remake a new data matrix which characters refer to the topologies of the source trees � � Baum, B. R. (1992) Taxon 41: 3 -10 Ragan, M. A. (1992) Mol. Phylo. Evol. 1: 53 -58 Sanderson. et al. (1998) TREE 13: 105 -109 16

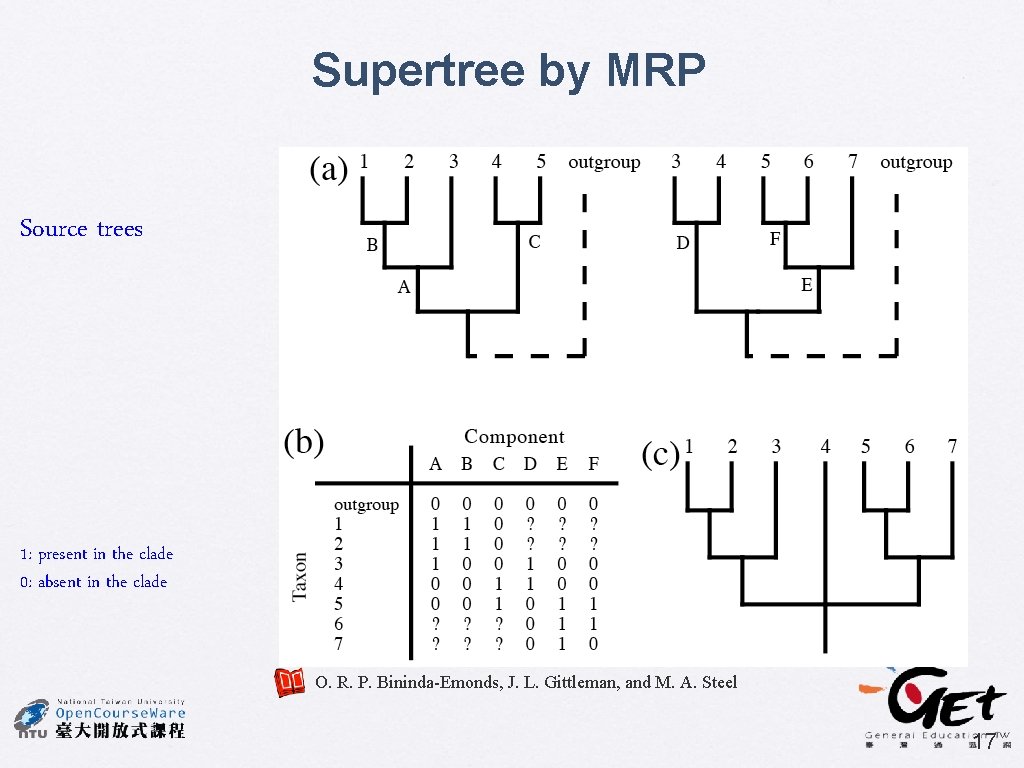
Supertree by MRP Source trees 1: present in the clade 0: absent in the clade O. R. P. Bininda-Emonds, J. L. Gittleman, and M. A. Steel 17

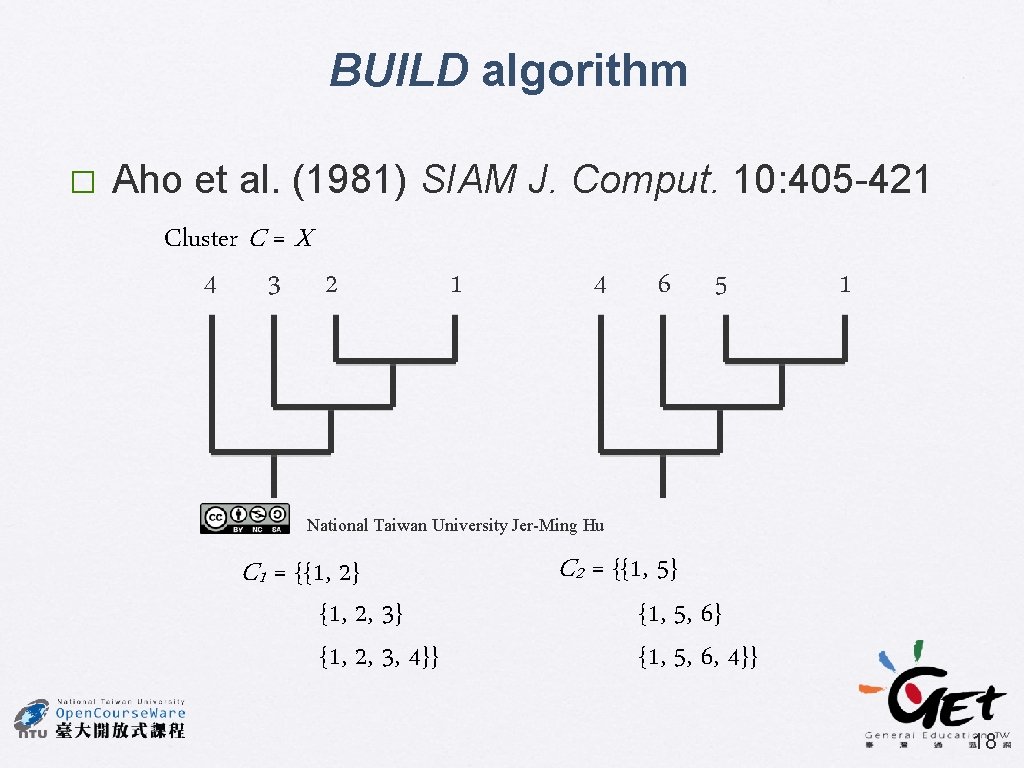
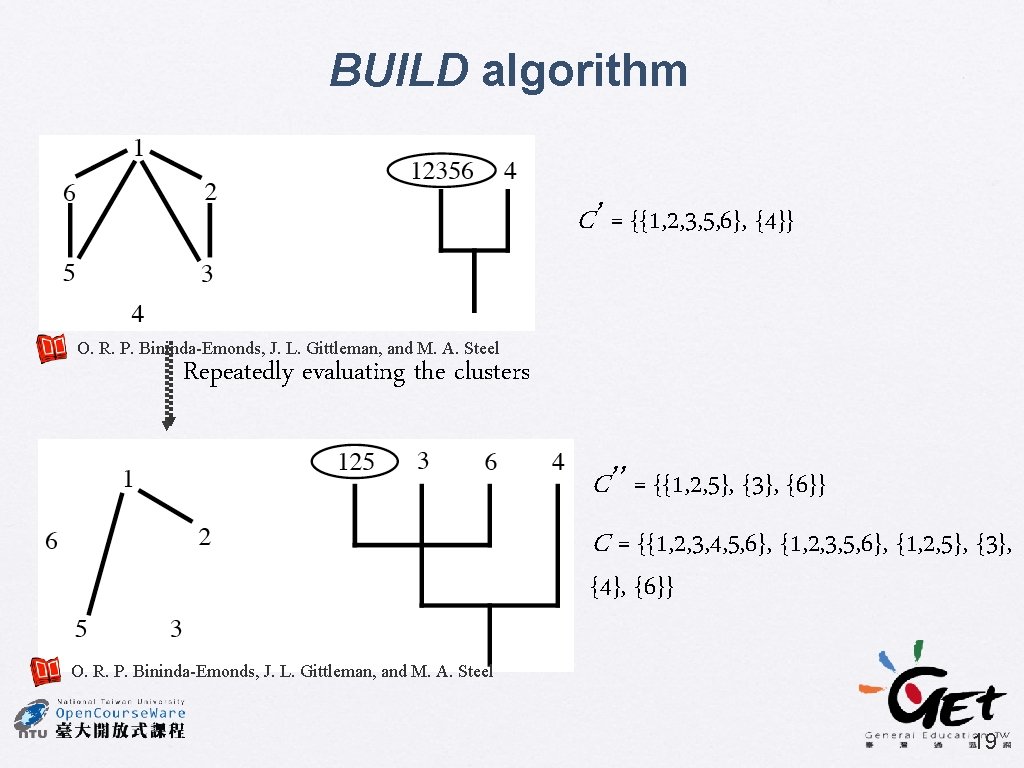
BUILD algorithm � Aho et al. (1981) SIAM J. Comput. 10: 405 -421 Cluster C = X 4 3 2 1 4 6 5 1 National Taiwan University Jer-Ming Hu C 1 = {{1, 2} {1, 2, 3, 4}} C 2 = {{1, 5} {1, 5, 6, 4}} 18

BUILD algorithm C’ = {{1, 2, 3, 5, 6}, {4}} O. R. P. Bininda-Emonds, J. L. Gittleman, and M. A. Steel Repeatedly evaluating the clusters C’’ = {{1, 2, 5}, {3}, {6}} C = {{1, 2, 3, 4, 5, 6}, {1, 2, 3, 5, 6}, {1, 2, 5}, {3}, {4}, {6}} O. R. P. Bininda-Emonds, J. L. Gittleman, and M. A. Steel 19

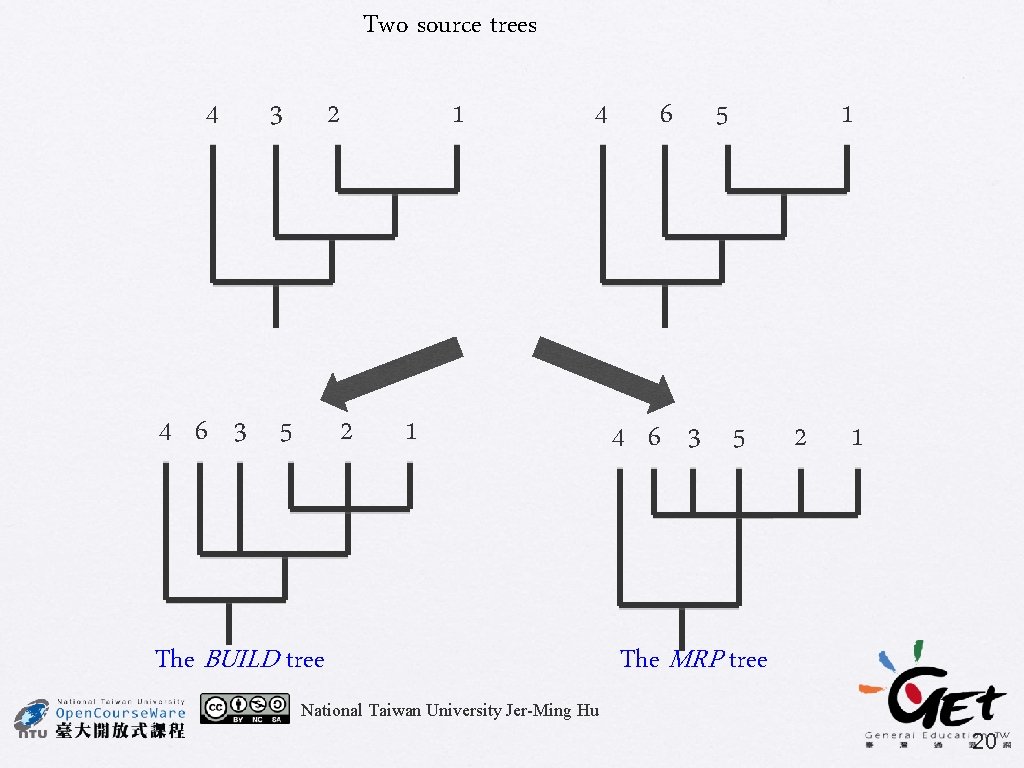
Two source trees 4 3 2 4 6 3 5 1 2 4 1 The BUILD tree 6 5 4 6 3 5 1 2 1 The MRP tree National Taiwan University Jer-Ming Hu 20

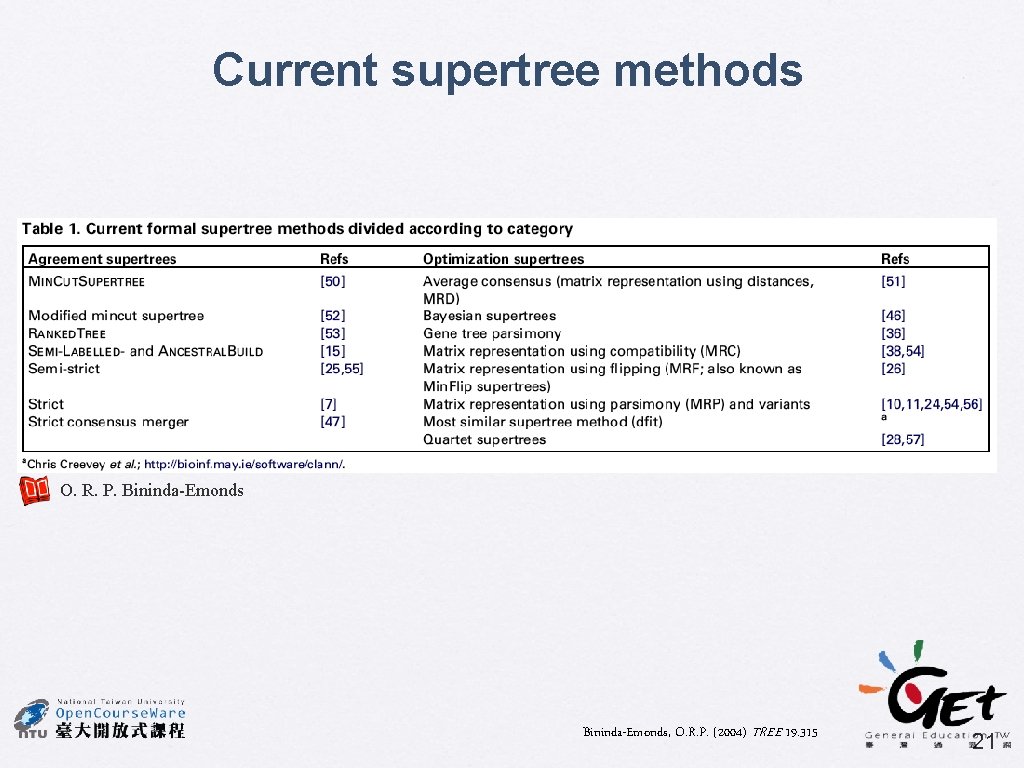
Current supertree methods O. R. P. Bininda-Emonds, O. R. P. (2004) TREE 19. 315 21

O. R. P. Bininda-Emonds 22

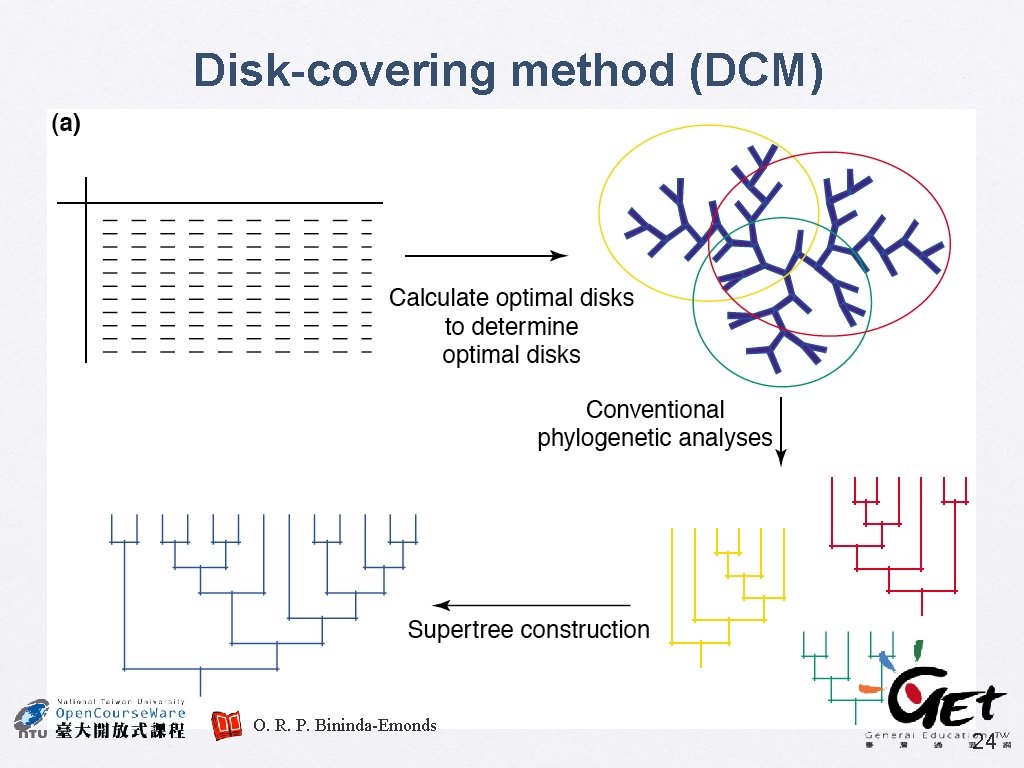
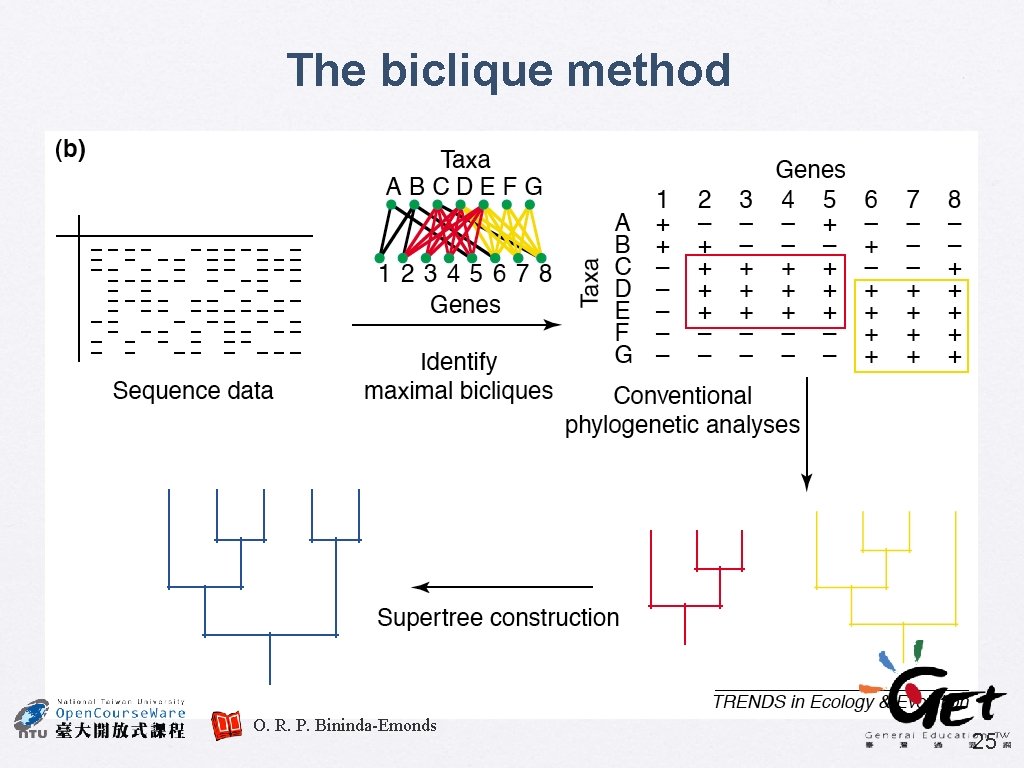
Supertree: the future � A divide-and-conquer approach can be applied, to decompose very large phylogenetic problem into many smaller subproblems, and the solutions to which are combined to derive the global answer Similar approaches have been used in phylogenetics, e. g. compartmentalization and quartet puzzling Two new approaches � � • • Disk-covering methods (DCMs) The biclique method Bininda-Emonds, O. R. P. (2004) TREE 19. 315 23

Disk-covering method (DCM) O. R. P. Bininda-Emonds 24

The biclique method O. R. P. Bininda-Emonds 25

Critics of supertree approach n It does not take sequence info into account n The dataset is non-independent n MRP favors larger source trees because they contribute more characters Some direct methods cannot incorporate support values into the source trees n 26

Programs available � supertree � � � Rod Page http: //darwin. zoology. gla. ac. uk/~rpage/supertre e/ Super. Tree Salamin et al. (2002) � http: //www 2. unil. ch/phylo/bioinformatics/supertr ee. html Supertrees related � Olaf R. P. Bininda-Emonds � http: //www. molekularesystematik. unioldenburg. de/en/34011. html � 27

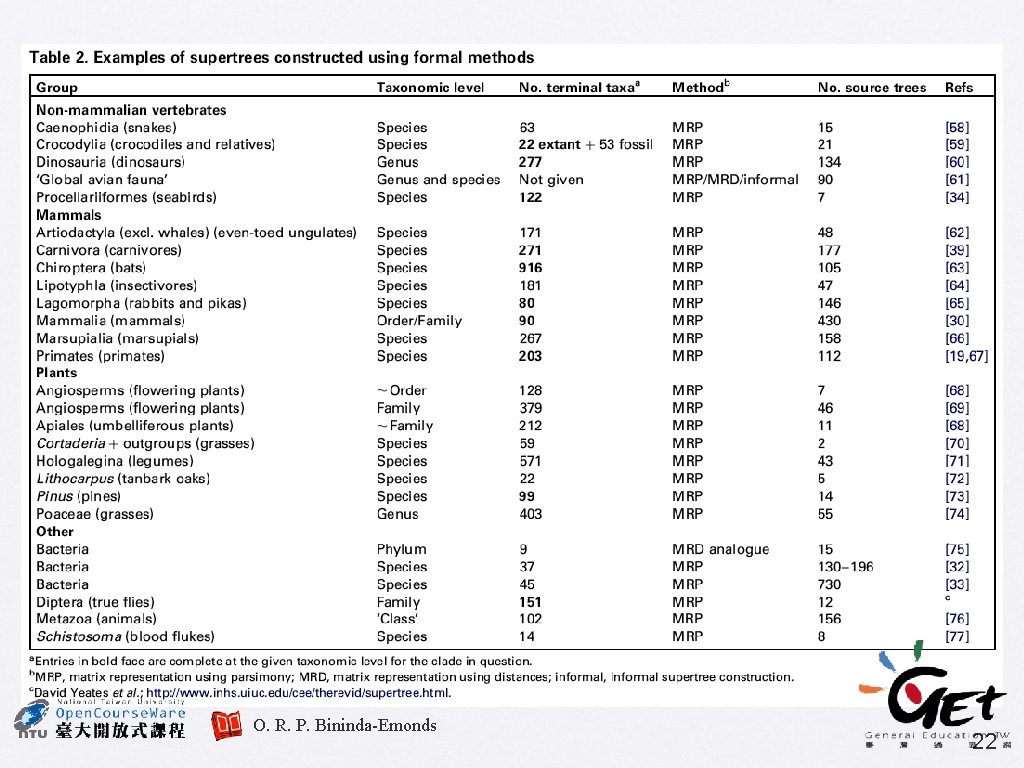
. . . okay, let’s see some real data � Supertree of legumes related to Astragalus � � Supertree of prokaryotes � � Sanderson, M. J. et al. (1998) TREE 13: 105 -109 Daubin, V. et al. (2002) Genome Res. 12: 1080 Supertree of the angiosperms � Davies, T. J. et al. (2004) PNAS 101: 1904 -1909 28

M. J. Sanderson, A. Purvis, and C. Henze Bold faces: taxa in common M. J. Sanderson, A. Purvis, and C. Henze 29

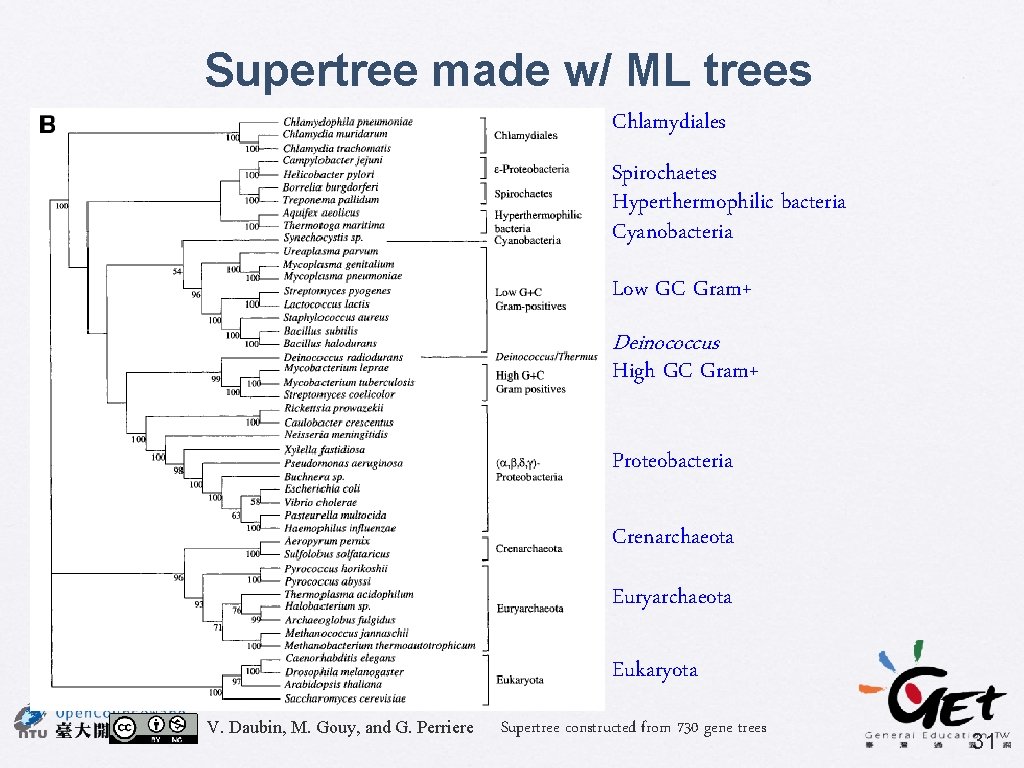
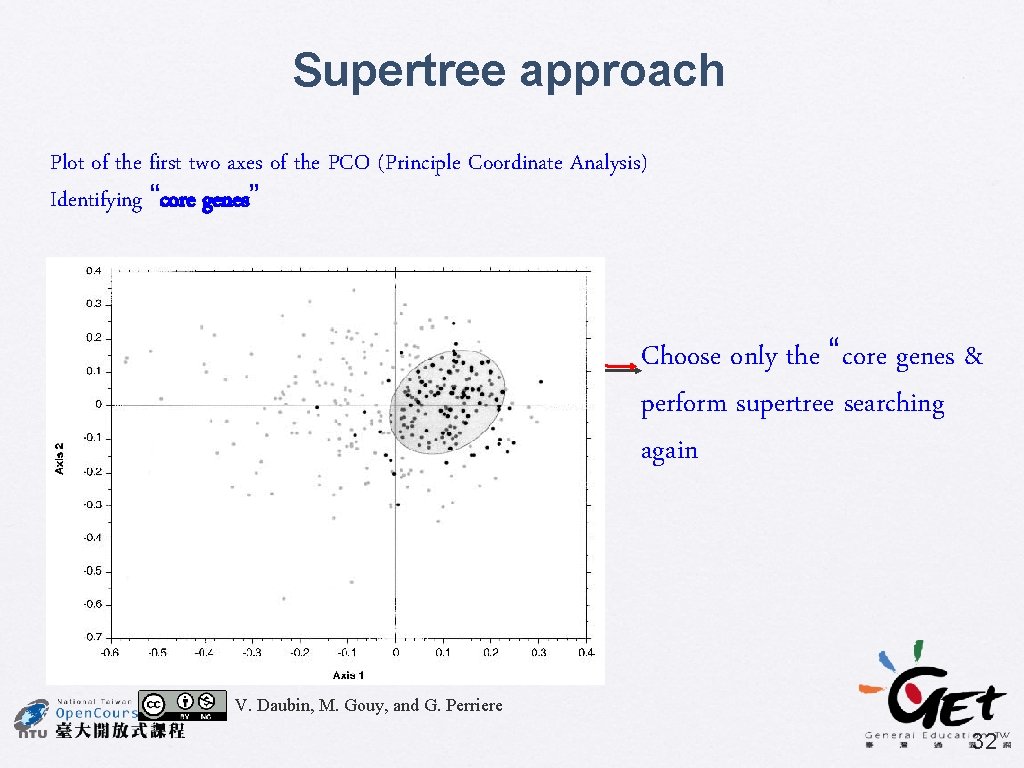
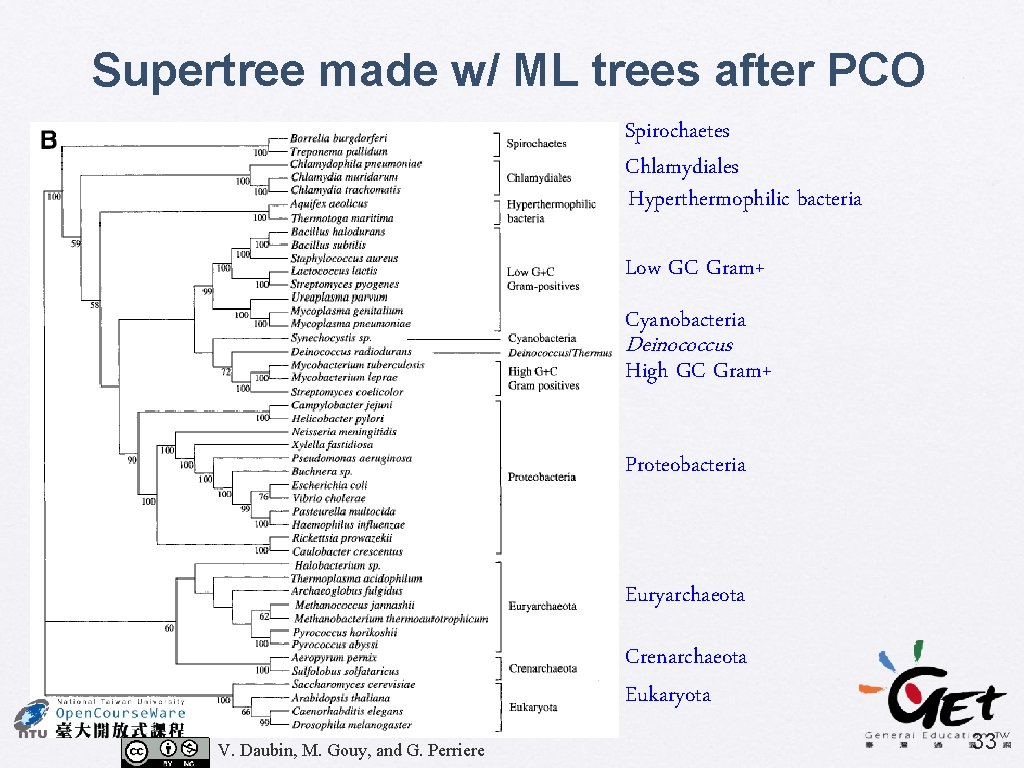
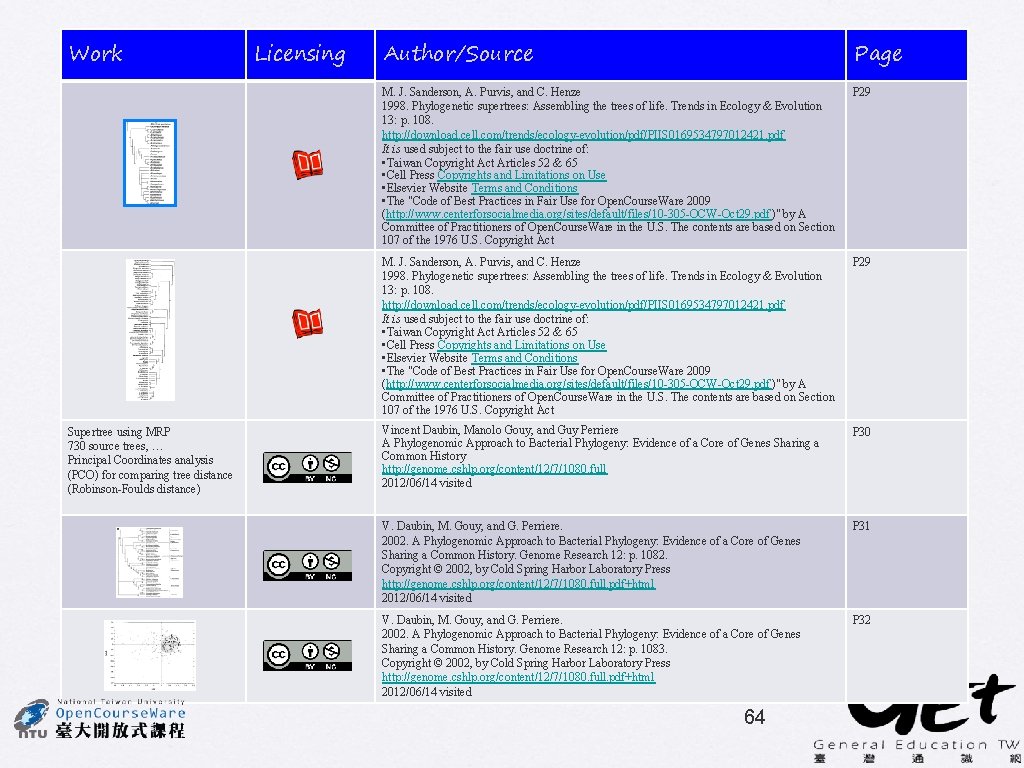
Genome Research (2002) 12: 1080 -1090 A Phylogenomic Approach to Bacterial Phylogeny: Evidence of a Core of Genes Sharing a Common History Vincent Daubin, Manolo Gouy, and Guy Perriere • Supertree using MRP • 730 source trees, include 45 taxa • Bootstrap % used as character weights • BIONJ + gamma • Identify the core of genes • Principal Coordinates analysis (PCO) for comparing tree distance (Robinson-Foulds distance) V. Daubin, M. Gouy, and G. Perriere 30

Supertree made w/ ML trees Chlamydiales Spirochaetes Hyperthermophilic bacteria Cyanobacteria Low GC Gram+ Deinococcus High GC Gram+ Proteobacteria Crenarchaeota Euryarchaeota Eukaryota V. Daubin, M. Gouy, and G. Perriere Supertree constructed from 730 gene trees 31

Supertree approach Plot of the first two axes of the PCO (Principle Coordinate Analysis) Identifying “core genes” Choose only the “core genes & perform supertree searching again V. Daubin, M. Gouy, and G. Perriere 32

Supertree made w/ ML trees after PCO Spirochaetes Chlamydiales Hyperthermophilic bacteria Low GC Gram+ Cyanobacteria Deinococcus High GC Gram+ Proteobacteria Euryarchaeota Crenarchaeota Eukaryota V. Daubin, M. Gouy, and G. Perriere 33

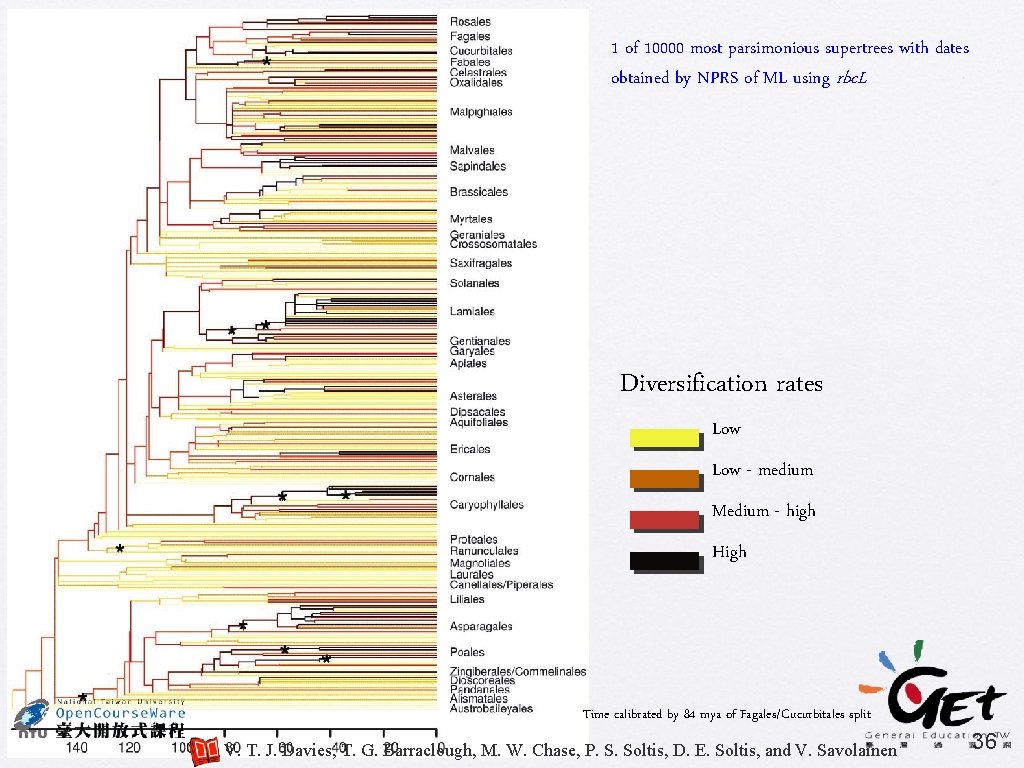
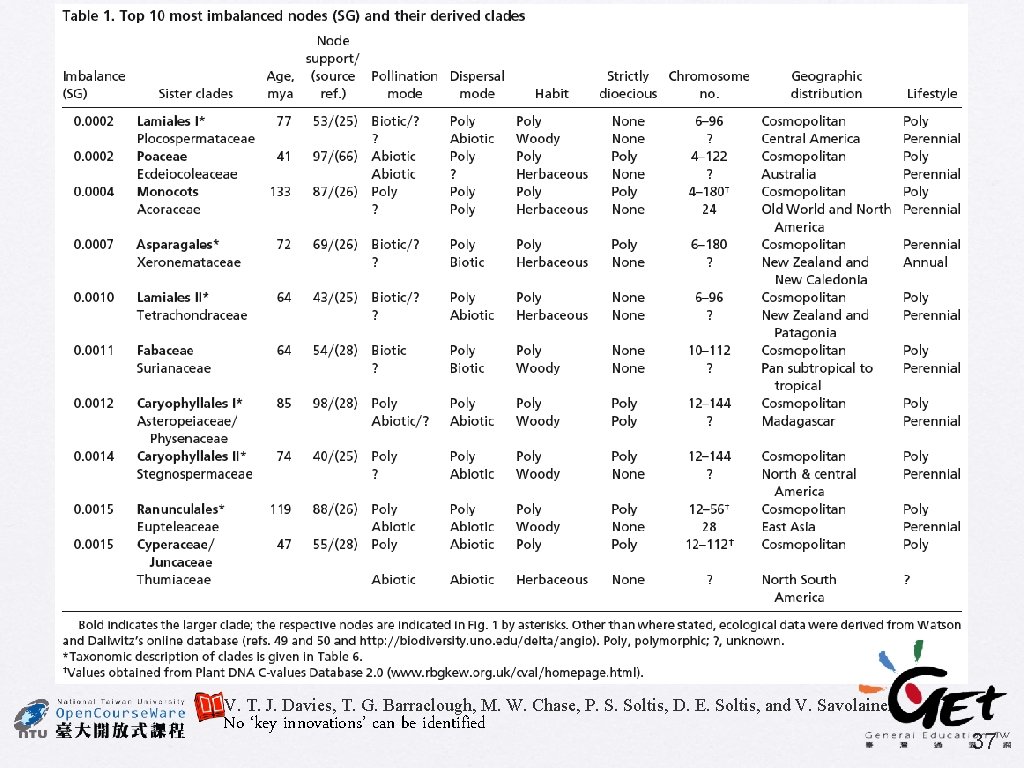
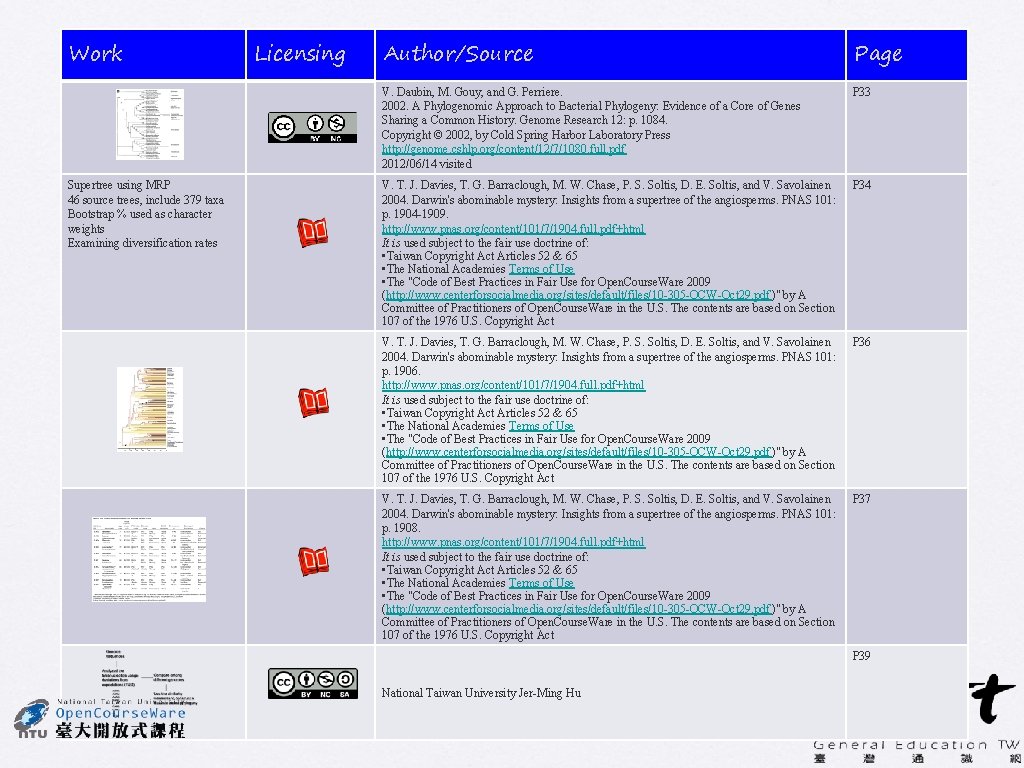
PNAS (2004) 101: 1904 -1909 Darwin's abominable mystery: Insights from a supertree of the angiosperms T. Johathan Davies, Timothy G. Barraclough, Mark W. Chase, Pamela S. Soltis, Douglas E. Soltis, and Vincent Savolainen • Supertree using MRP • 46 source trees, include 379 taxa • Bootstrap % used as character weights • Examining diversification rates V. T. J. Davies, T. G. Barraclough, M. W. Chase, P. S. Soltis, D. E. Soltis, and V. Savolainen 34

PNAS (2004) 101: 1904 -1909 Darwin's abominable mystery: Insights from a supertree of the angiosperms T. Johathan Davies, Timothy G. Barraclough, Mark W. Chase, Pamela S. Soltis, Douglas E. Soltis, and Vincent Savolainen Supporting Table 2 35

1 of 10000 most parsimonious supertrees with dates obtained by NPRS of ML using rbc. L Diversification rates Low - medium Medium - high High Time calibrated by 84 mya of Fagales/Cucurbitales split V. T. J. Davies, T. G. Barraclough, M. W. Chase, P. S. Soltis, D. E. Soltis, and V. Savolainen 36

V. T. J. Davies, T. G. Barraclough, M. W. Chase, P. S. Soltis, D. E. Soltis, and V. Savolainen No ‘key innovations’ can be identified 37

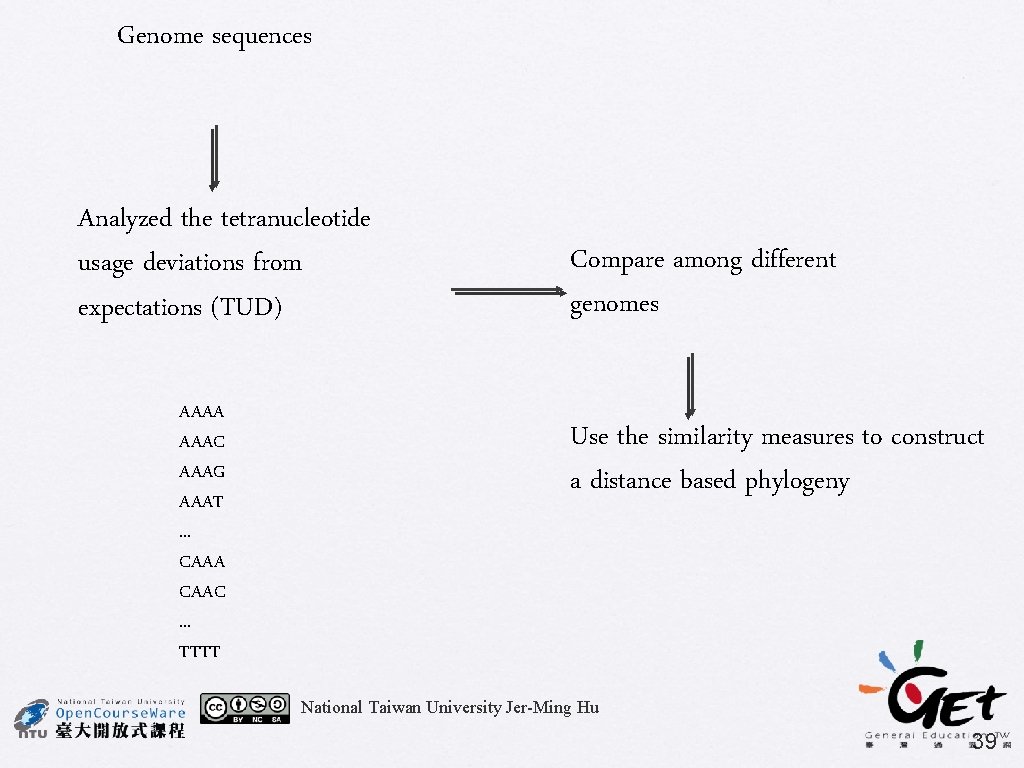
Phylogenomics using whole-genome features � Distribution of sequence strings � � � Transformation of the frequencies of small oligonucleotides (up to eight) or oligopeptides (up to six) into distances and used for phylogenetic reconstruction Gene order methods Gene content methods 38

Genome sequences Analyzed the tetranucleotide usage deviations from expectations (TUD) AAAA AAAC AAAG AAAT. . . CAAA CAAC. . . TTTT Compare among different genomes Use the similarity measures to construct a distance based phylogeny National Taiwan University Jer-Ming Hu 39

Potential problems using sequence strings � � The phylogeneic information of sequence strings saturates rapidly No models of evolution is proposed for such distribution of oligonucleotide or oligopeptide frequencies 40

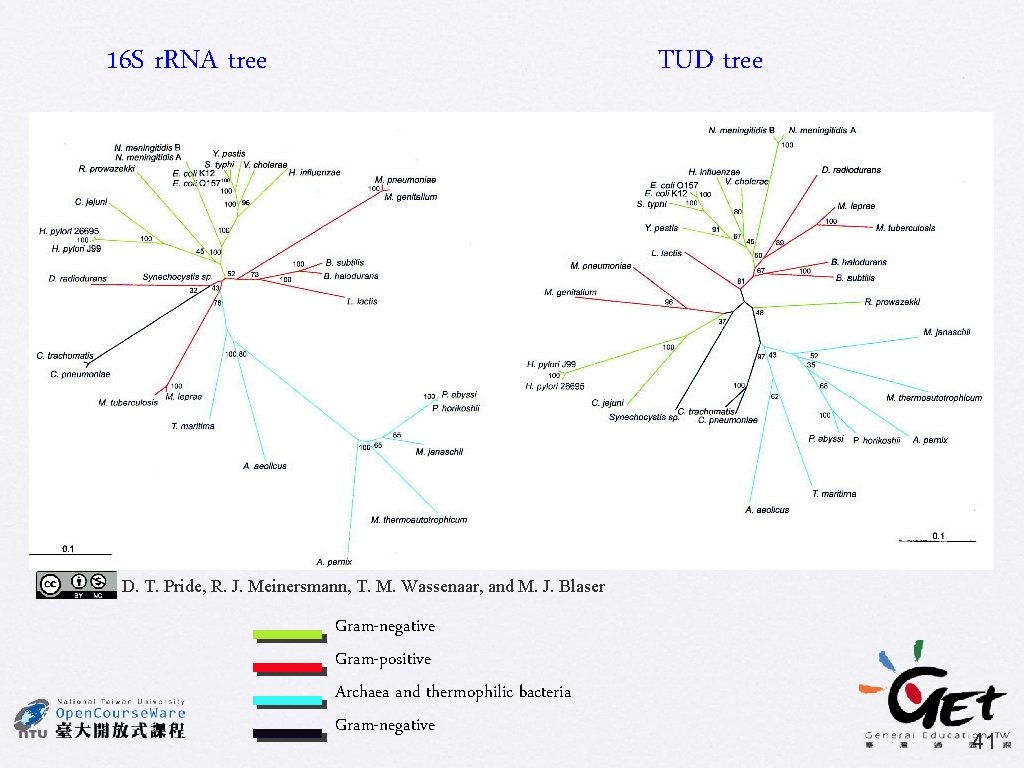
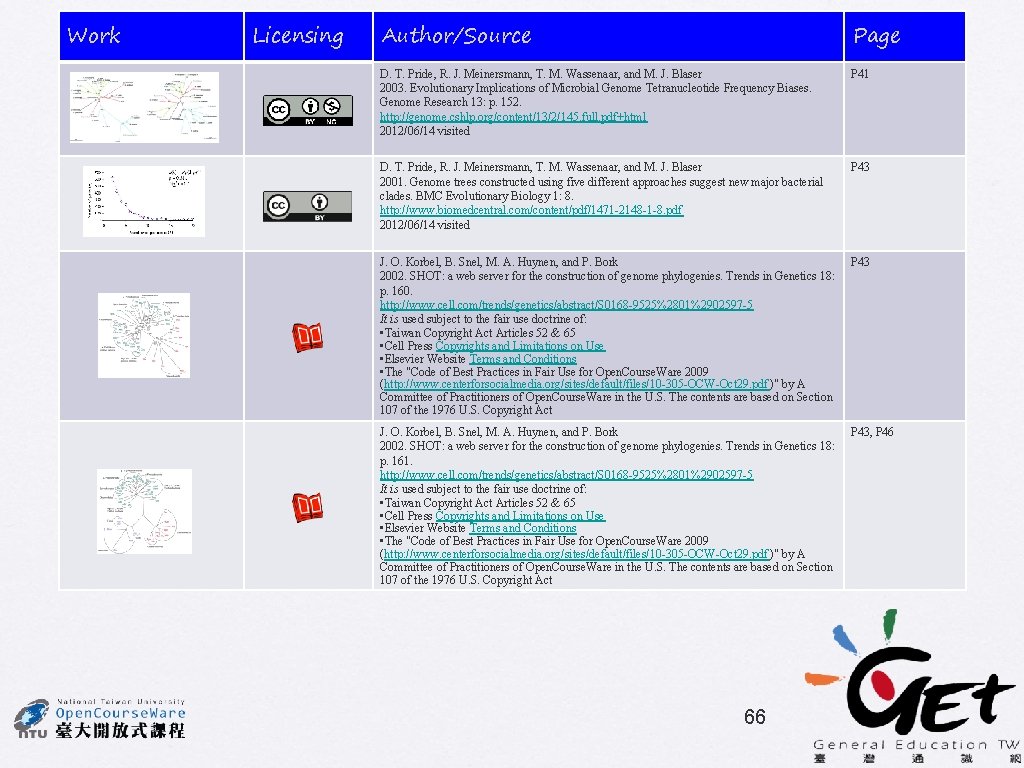
16 S r. RNA tree TUD tree D. T. Pride, R. J. Meinersmann, T. M. Wassenaar, and M. J. Blaser Gram-negative Gram-positive Archaea and thermophilic bacteria Gram-negative 41

Gene order method � � � Information of genome organization Distances of two genomes can be computed to minimize the numbers of inversions, transpositions, insertions, and deletions Distances can also be computed for the minimum number of pairs of genes next to each other in one of the two genomes, but not in the other (“break-point distance”) 42

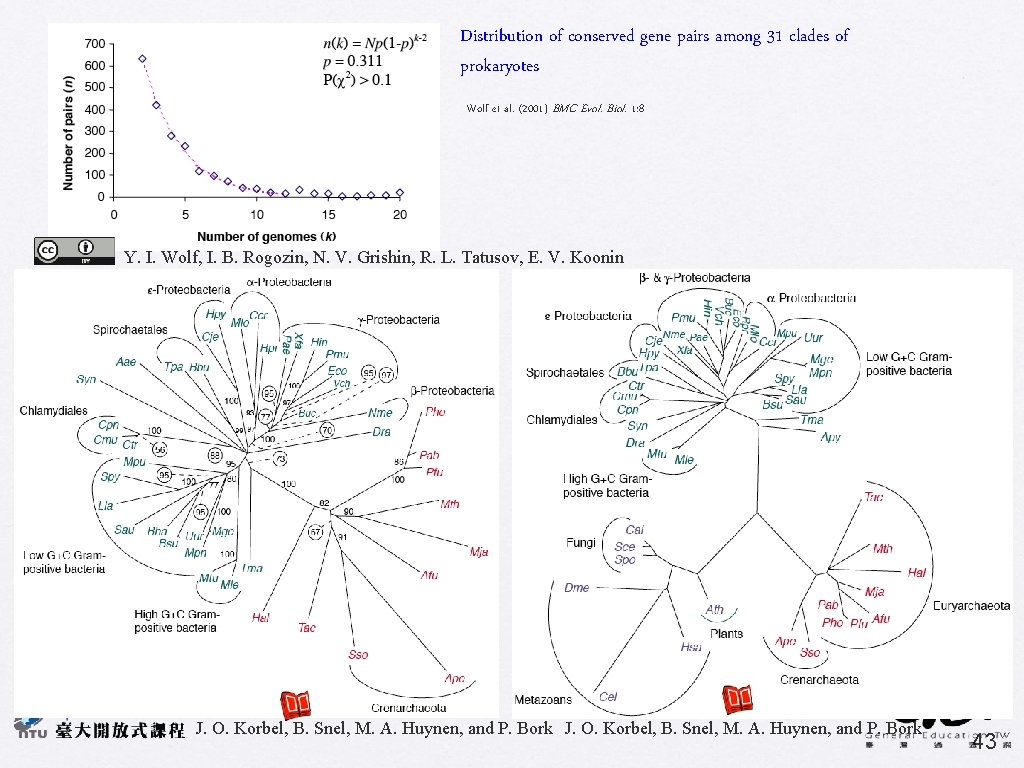
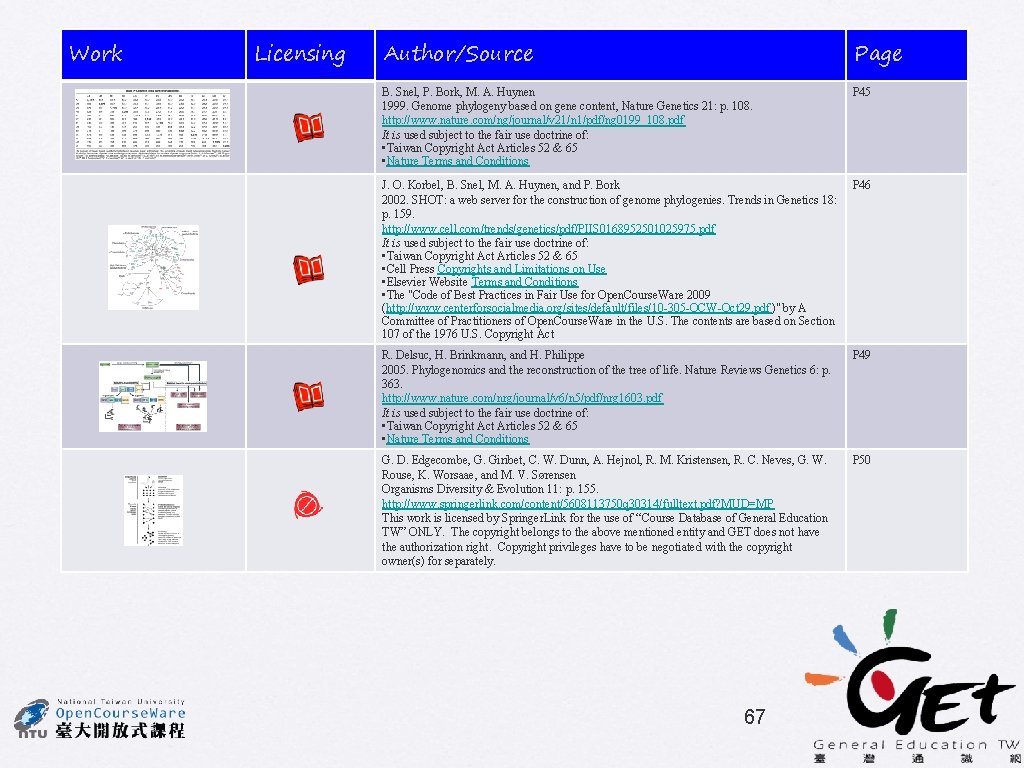
Distribution of conserved gene pairs among 31 clades of prokaryotes Wolf et al. (2001) BMC Evol. Biol. 1: 8 Y. I. Wolf, I. B. Rogozin, N. V. Grishin, R. L. Tatusov, E. V. Koonin Gene-order phylogeny ss r. RNA phylogeny J. O. Korbel, B. Snel, M. A. Huynen, and P. Bork 43

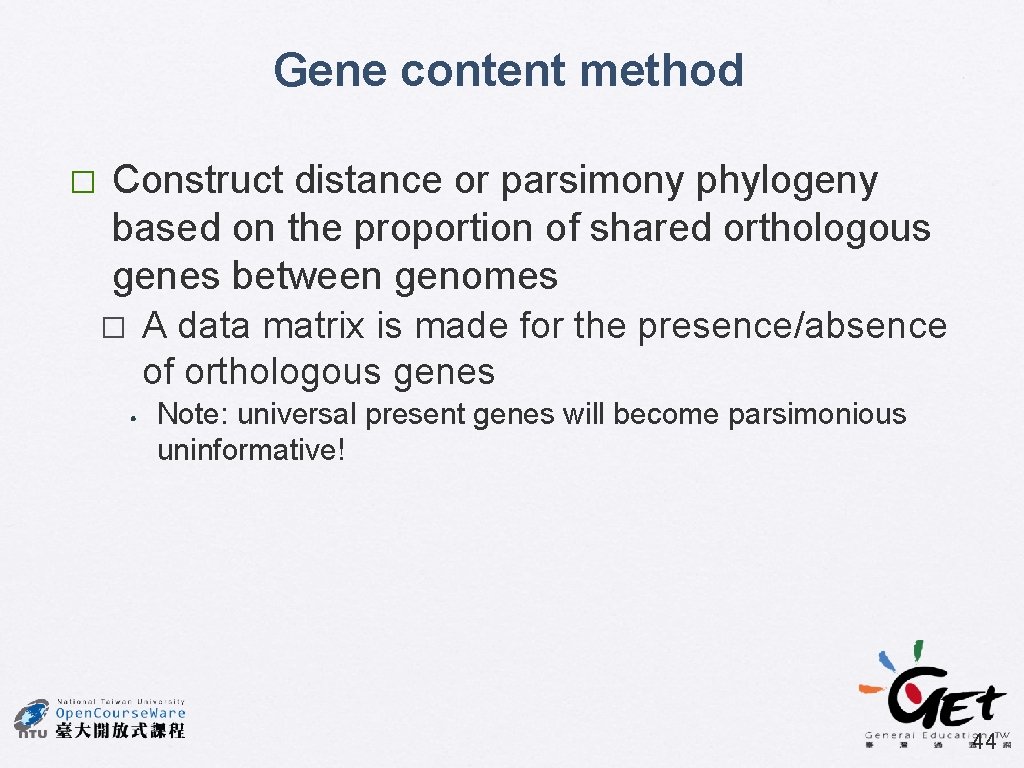
Gene content method � Construct distance or parsimony phylogeny based on the proportion of shared orthologous genes between genomes A data matrix is made for the presence/absence of orthologous genes � • Note: universal present genes will become parsimonious uninformative! 44

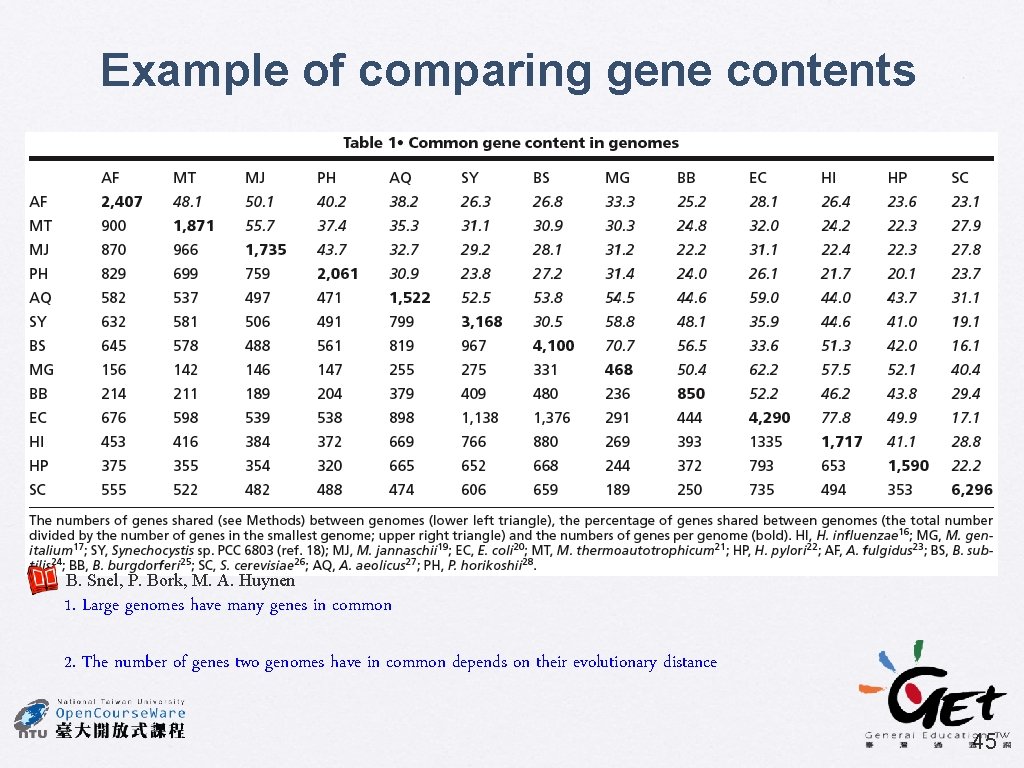
Example of comparing gene contents B. Snel, P. Bork, M. A. Huynen 1. Large genomes have many genes in common 2. The number of genes two genomes have in common depends on their evolutionary distance 45

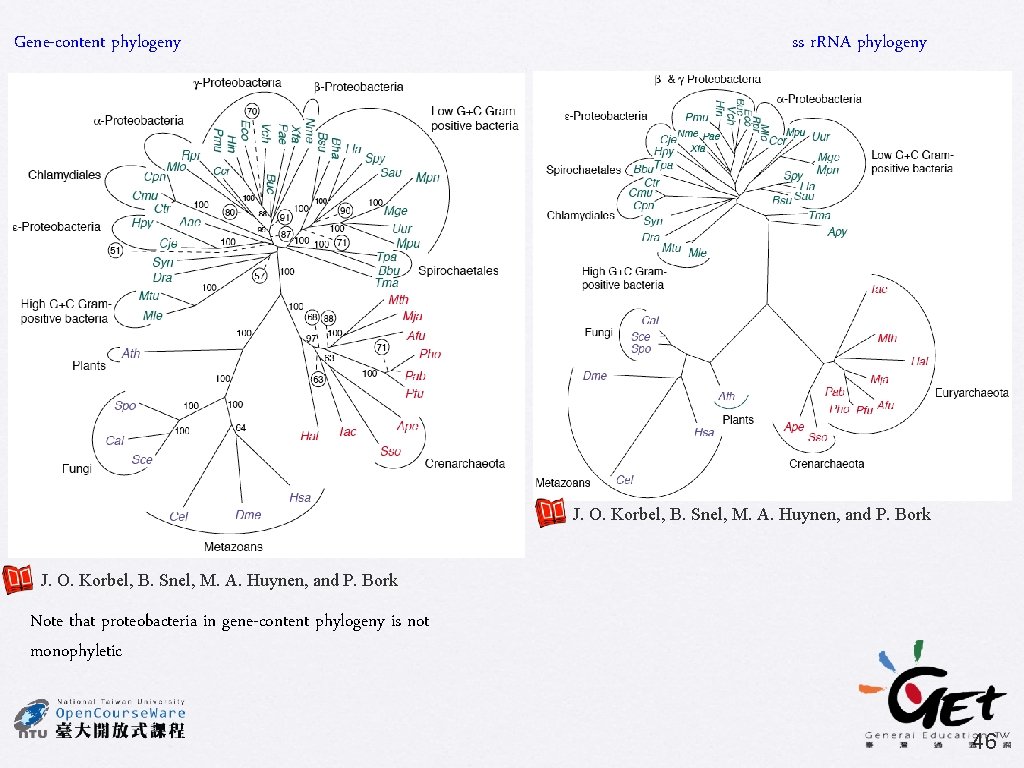
Gene-content phylogeny ss r. RNA phylogeny J. O. Korbel, B. Snel, M. A. Huynen, and P. Bork Note that proteobacteria in gene-content phylogeny is not monophyletic 46

Problems using gene-content method � Big/small genome attraction � � Artificial grouping of unrelated species with small genomes, e. g. Mycoplasma, Buchnera, Chlamydia, or Rickettsia. Phylogenetic positions of some taxa do not agree with previous studies � Halobacterium never clusters with Methanosarcina. This is probably due to too many HGTs 47

Programs available � SHOT � � A web server for the construction of genome phylogenies http: //coot. embl. de/~korbel/SHOT/ 48

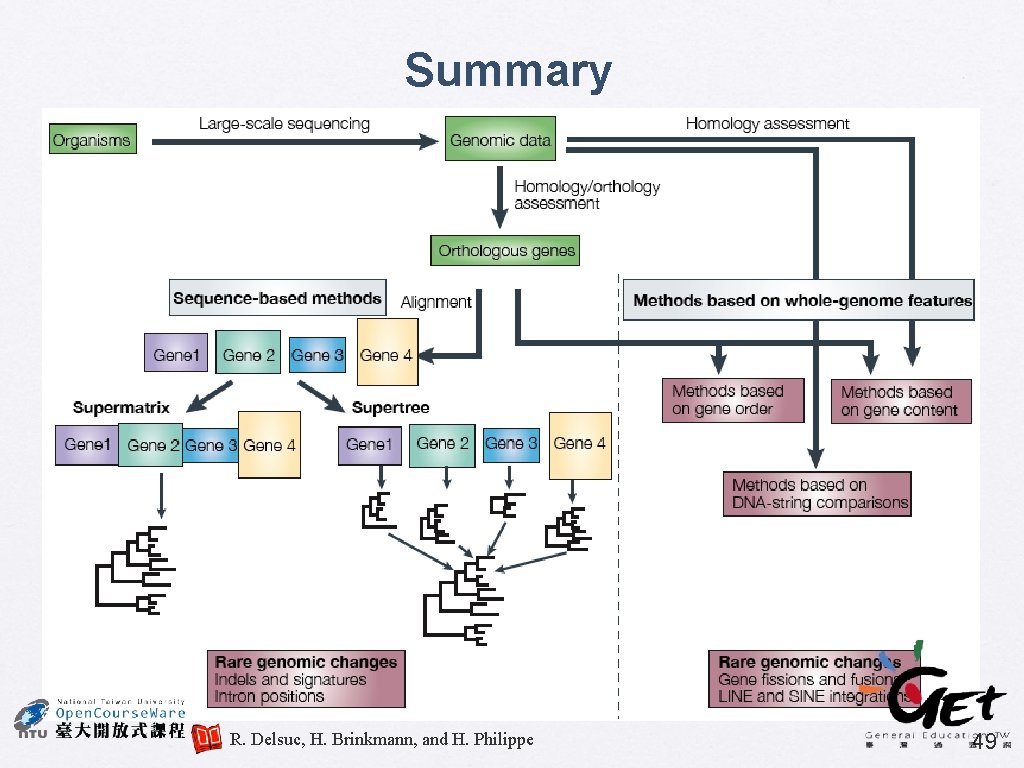
Summary R. Delsuc, H. Brinkmann, and H. Philippe 49

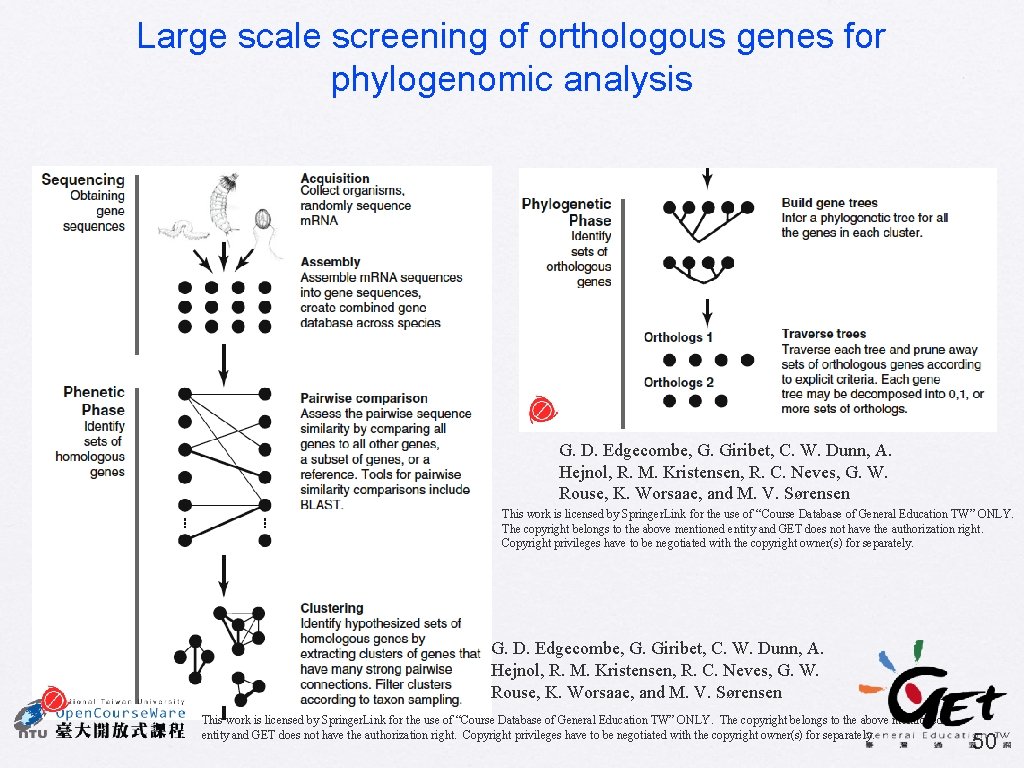
Large scale screening of orthologous genes for phylogenomic analysis G. D. Edgecombe, G. Giribet, C. W. Dunn, A. Hejnol, R. M. Kristensen, R. C. Neves, G. W. Rouse, K. Worsaae, and M. V. Sørensen This work is licensed by Springer. Link for the use of “Course Database of General Education TW” ONLY. The copyright belongs to the above mentioned entity and GET does not have the authorization right. Copyright privileges have to be negotiated with the copyright owner(s) for separately. 50

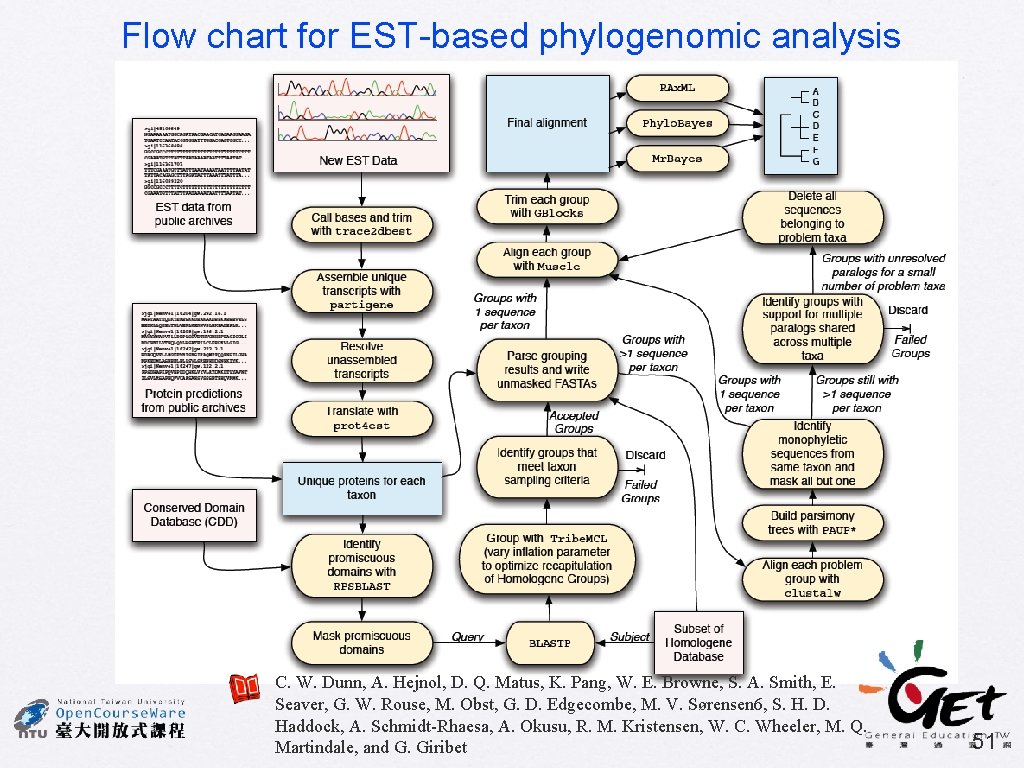
Flow chart for EST-based phylogenomic analysis C. W. Dunn, A. Hejnol, D. Q. Matus, K. Pang, W. E. Browne, S. A. Smith, E. Seaver, G. W. Rouse, M. Obst, G. D. Edgecombe, M. V. Sørensen 6, S. H. D. Haddock, A. Schmidt-Rhaesa, A. Okusu, R. M. Kristensen, W. C. Wheeler, M. Q. Martindale, and G. Giribet 51

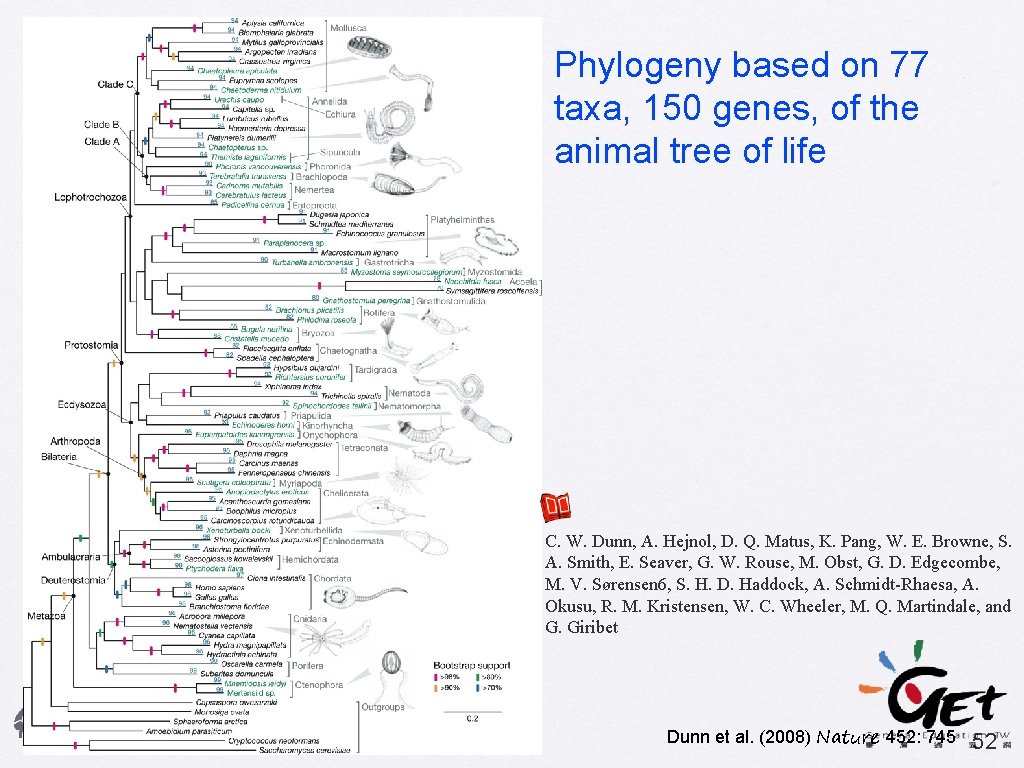
Phylogeny based on 77 taxa, 150 genes, of the animal tree of life C. W. Dunn, A. Hejnol, D. Q. Matus, K. Pang, W. E. Browne, S. A. Smith, E. Seaver, G. W. Rouse, M. Obst, G. D. Edgecombe, M. V. Sørensen 6, S. H. D. Haddock, A. Schmidt-Rhaesa, A. Okusu, R. M. Kristensen, W. C. Wheeler, M. Q. Martindale, and G. Giribet Dunn et al. (2008) Nature 452: 745 52

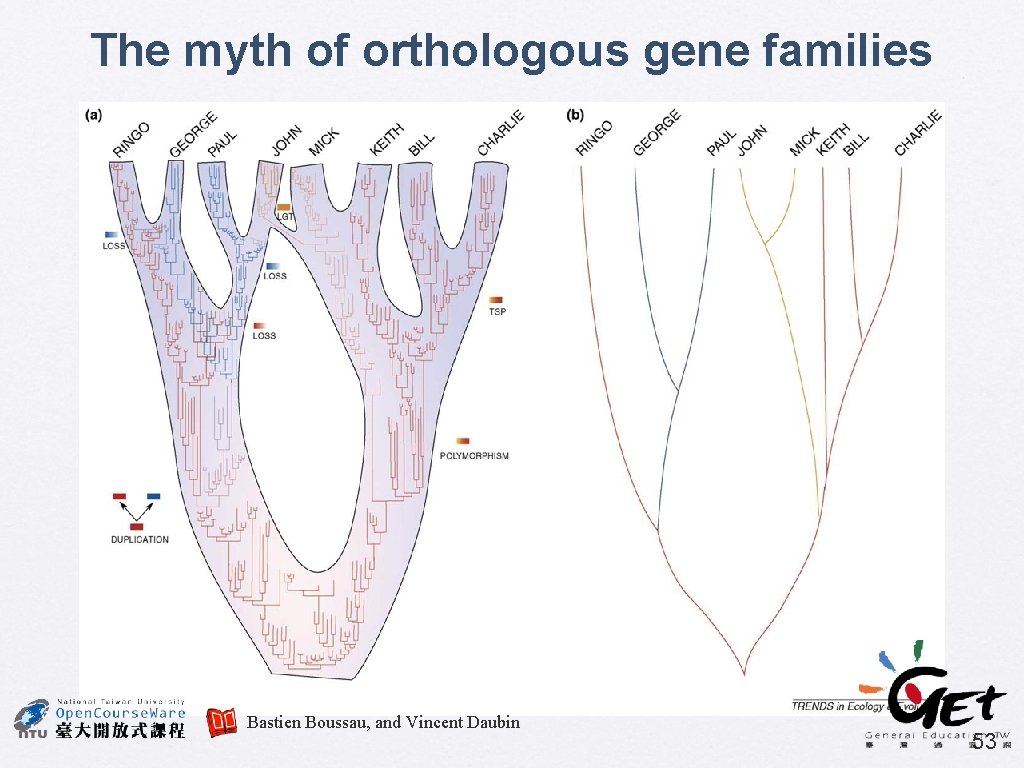
The myth of orthologous gene families Bastien Boussau, and Vincent Daubin 53

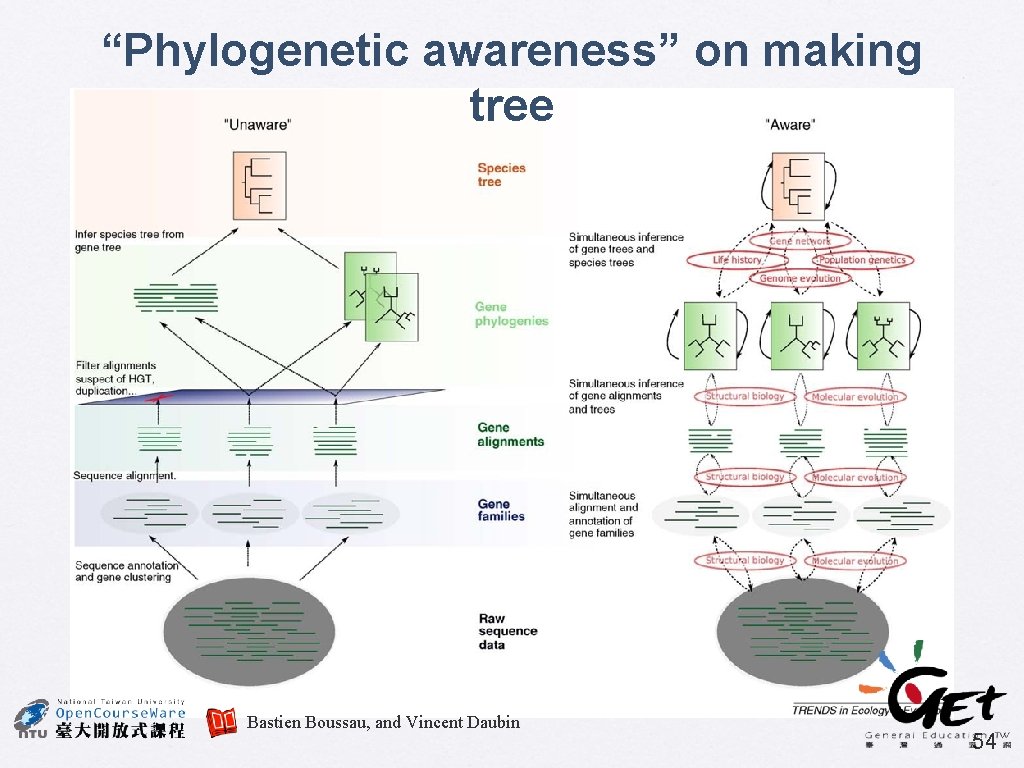
“Phylogenetic awareness” on making tree Bastien Boussau, and Vincent Daubin 54

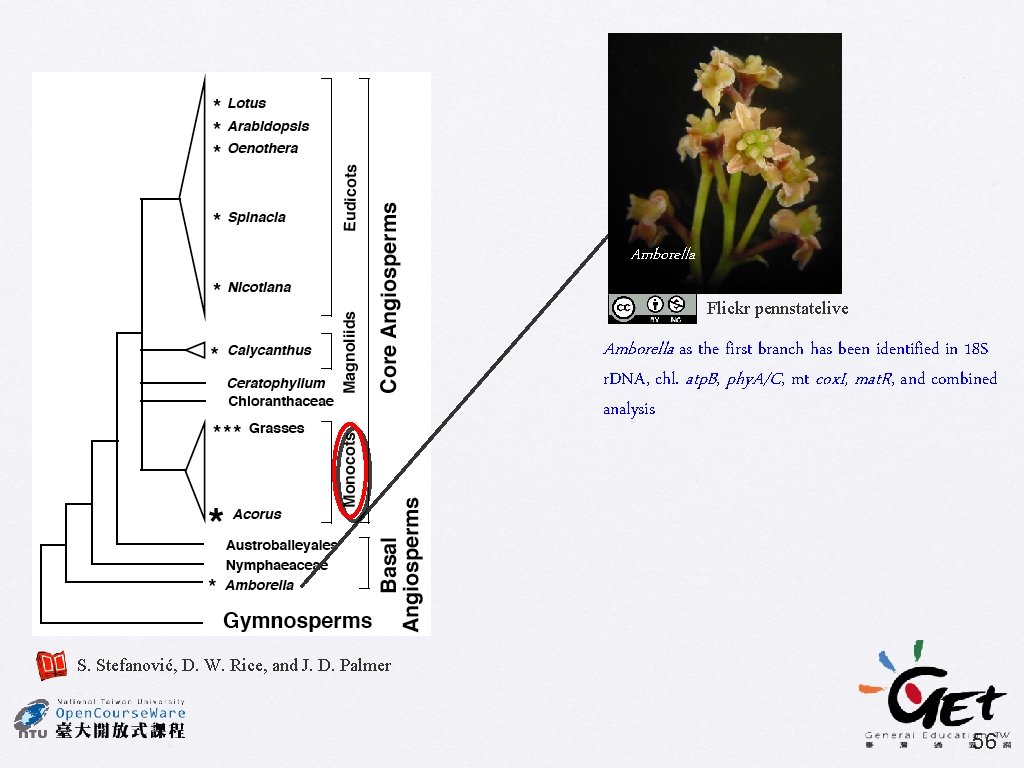
Problems of just using whole-genome data � Case study of seeking the root of angiosperms � Amborella or monocots? Flickr scott. zona Flickr adrien 2008 55

Amborella Flickr pennstatelive Amborella as the first branch has been identified in 18 S r. DNA, chl. atp. B, phy. A/C, mt cox. I, mat. R, and combined analysis S. Stefanović, D. W. Rice, and J. D. Palmer 56

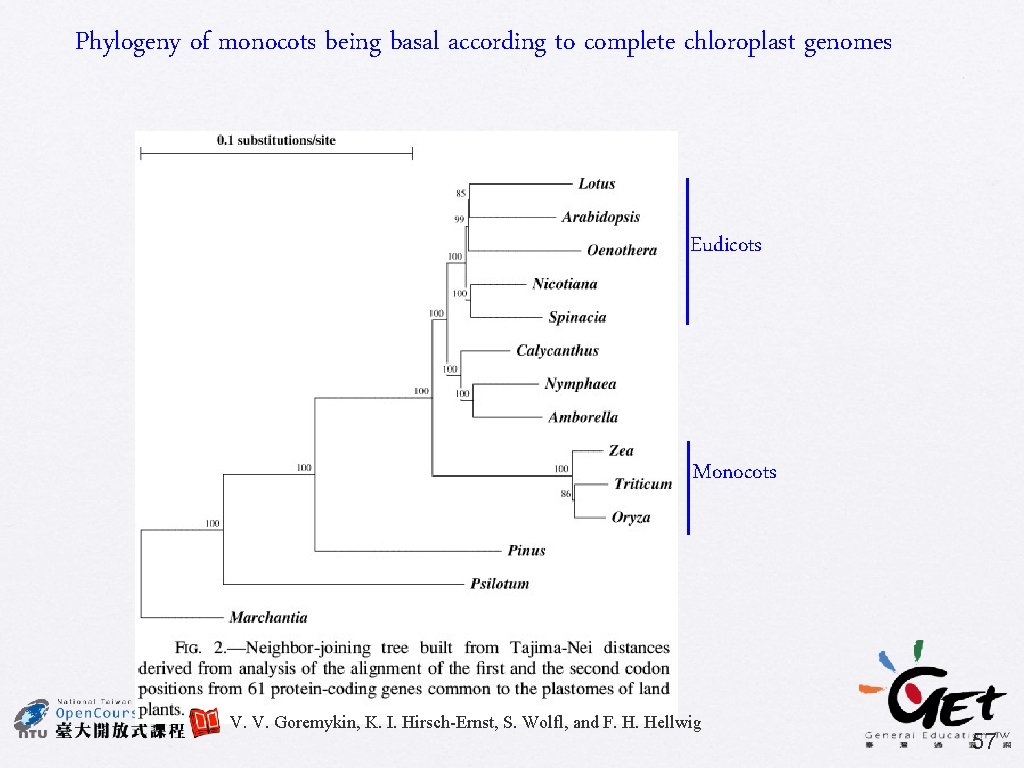
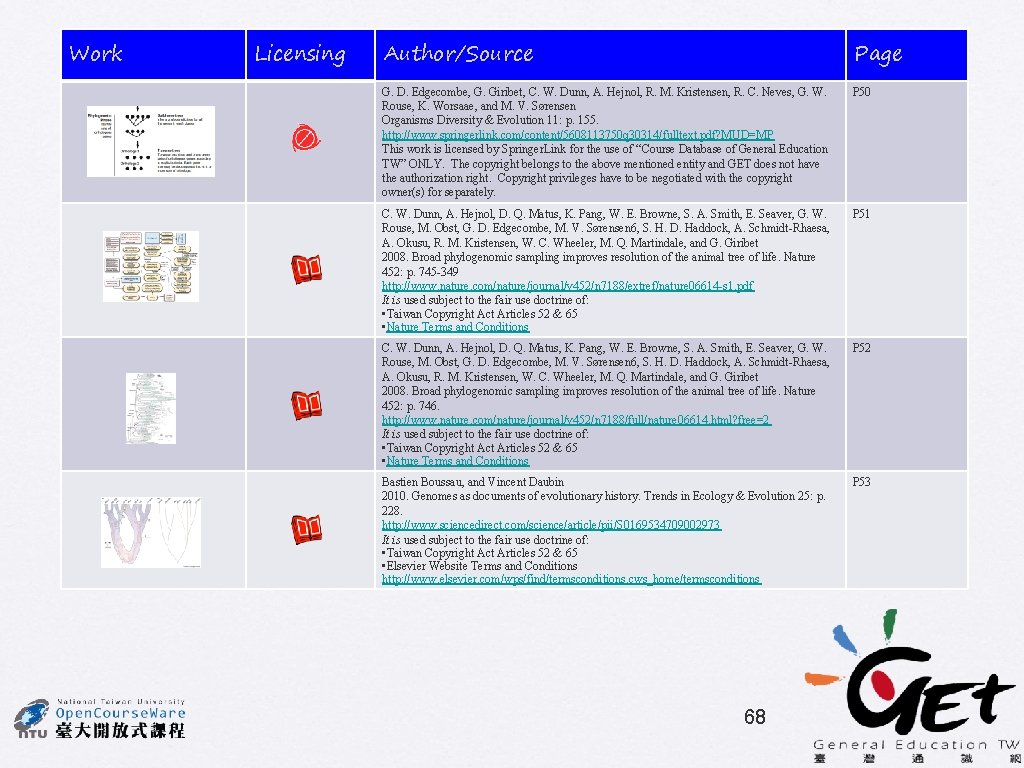
Phylogeny of monocots being basal according to complete chloroplast genomes Eudicots Monocots V. V. Goremykin, K. I. Hirsch-Ernst, S. Wolfl, and F. H. Hellwig 57

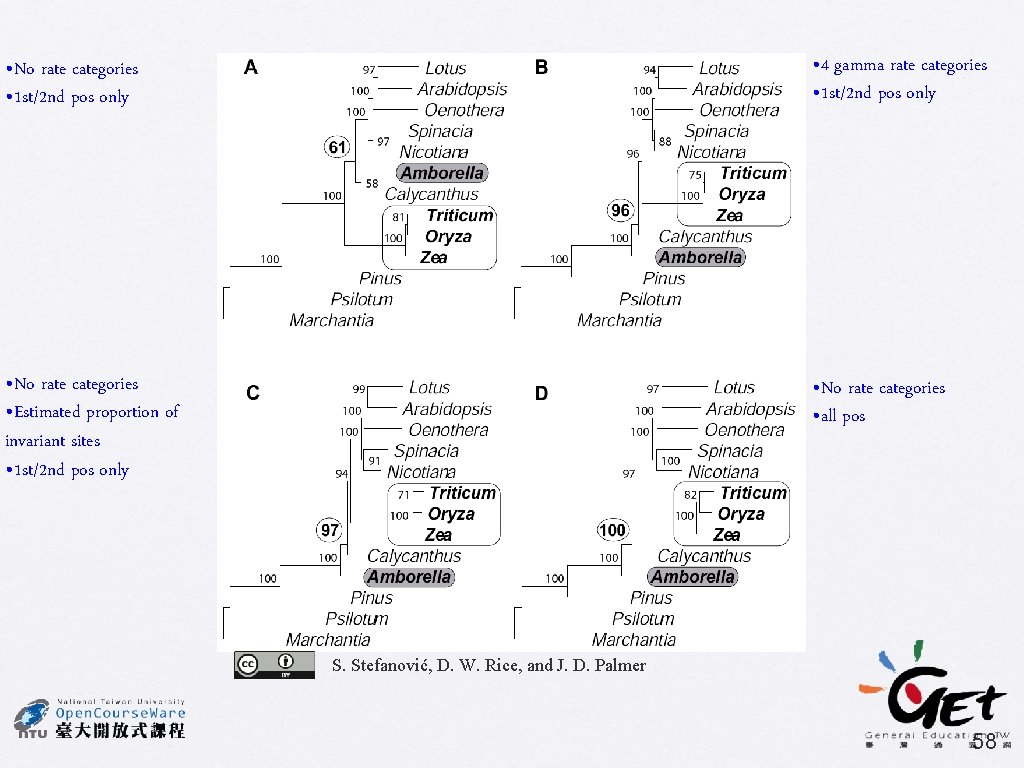
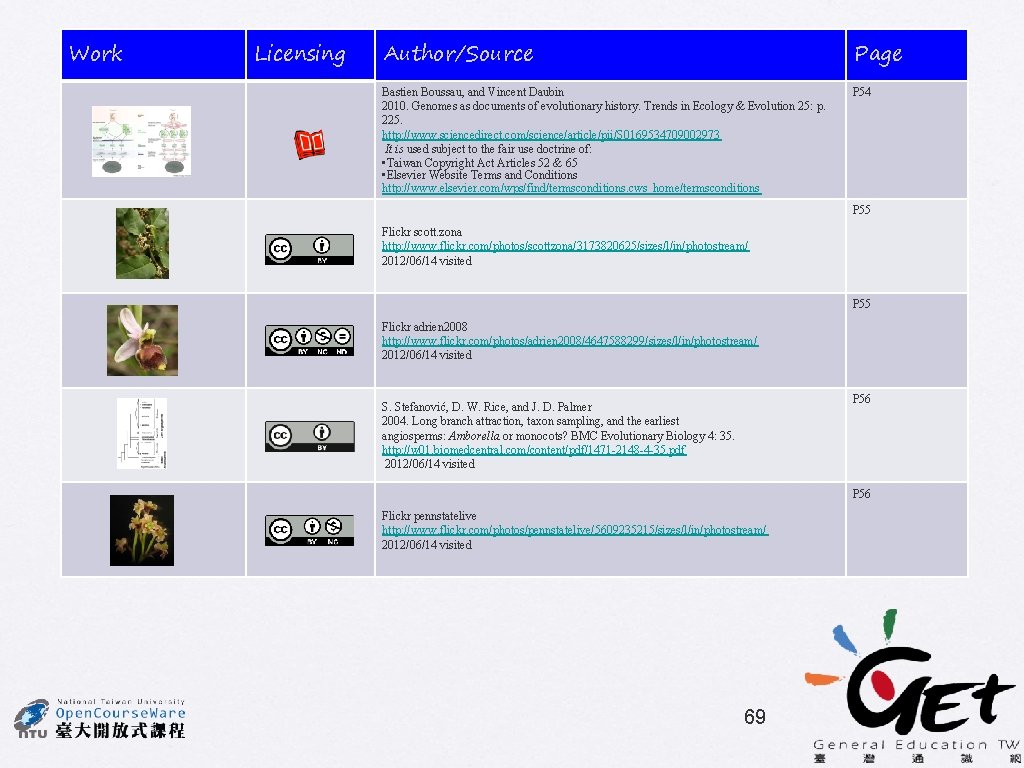
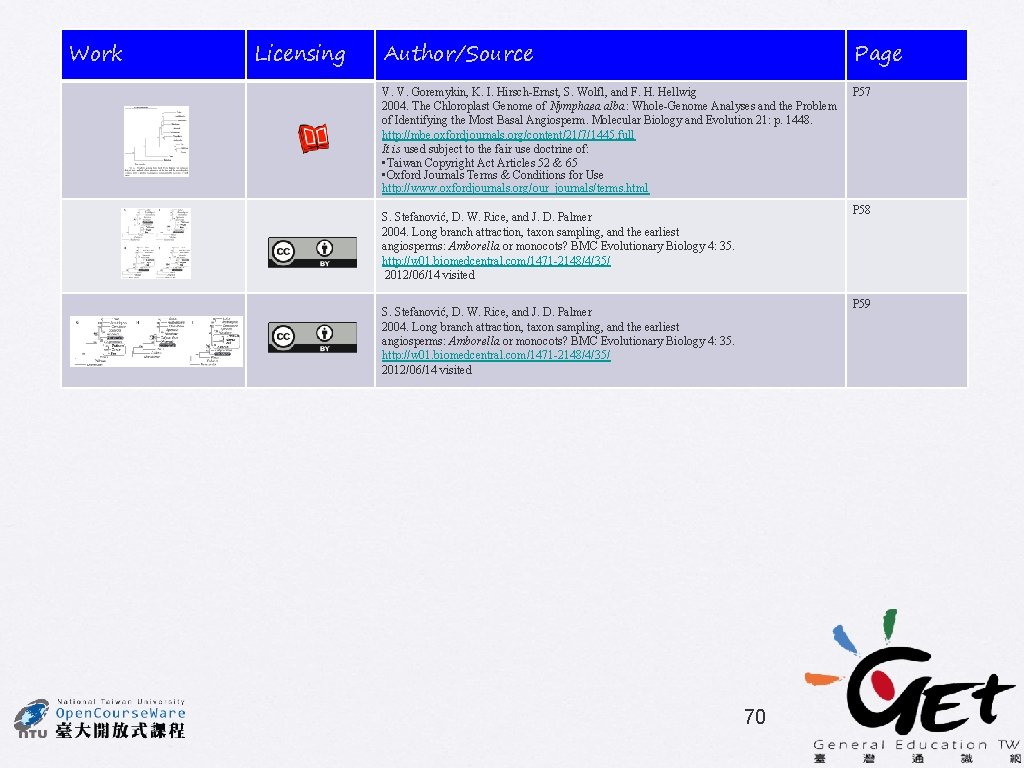
• No rate categories • 1 st/2 nd pos only • 4 gamma rate categories • 1 st/2 nd pos only • No rate categories • Estimated proportion of invariant sites • 1 st/2 nd pos only • No rate categories • all pos S. Stefanović, D. W. Rice, and J. D. Palmer 58

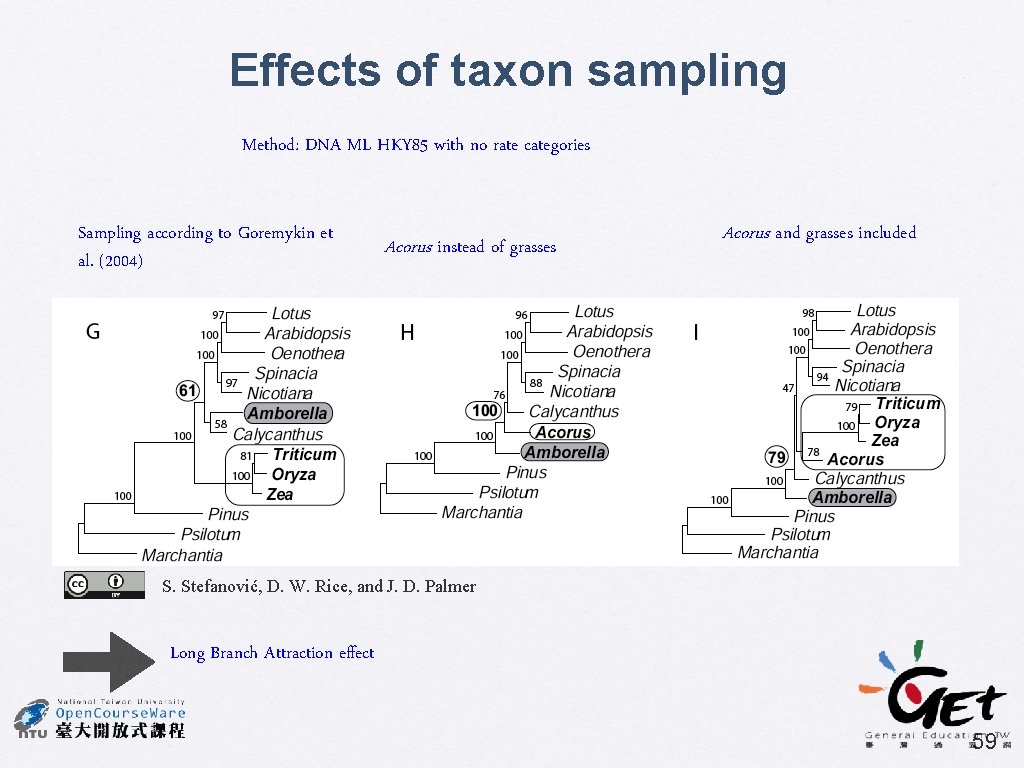
Effects of taxon sampling Method: DNA ML HKY 85 with no rate categories Sampling according to Goremykin et al. (2004) Acorus instead of grasses Acorus and grasses included S. Stefanović, D. W. Rice, and J. D. Palmer Long Branch Attraction effect 59

Copyright Declaration Work Licensing Author/Source Page P 8 National Taiwan University Jer-Ming Hu M. J. Sanderson and A. C. Driskell 2003. The challenge of constructing large phylogenetic trees. Trends in Plant Science 8: p. 374. http: //www. sciencedirect. com/science/article/pii/S 1360138503001651 It is used subject to the fair use doctrine of: • Taiwan Copyright Act Articles 52 & 65 • Elsevier Website Terms and Conditions http: //www. elsevier. com/wps/find/termsconditions. cws_home/termsconditions P 9 M. J. Sanderson and A. C. Driskell 2003. The challenge of constructing large phylogenetic trees. Trends in Plant Science 8: p. 375. http: //www. sciencedirect. com/science/article/pii/S 1360138503001651 It is used subject to the fair use doctrine of: • Taiwan Copyright Act Articles 52 & 65 • Elsevier Website Terms and Conditions http: //www. elsevier. com/wps/find/termsconditions. cws_home/termsconditions P 10 O. R. P. Bininda-Emonds 2004. The evolution of supertrees. Trends in Ecology & Evolution 19: p. 315. http: //www. sciencedirect. com/science/article/pii/S 0169534704000709 It is used subject to the fair use doctrine of: • Taiwan Copyright Act Articles 52 & 65 • Elsevier Website Terms and Conditions http: //www. elsevier. com/wps/find/termsconditions. cws_home/termsconditions P 12 60

Work Licensing Author/Source Page P 13 National Taiwan University Jer-Ming Hu M. J. Sanderson, A. Purvis, and C. Henze 1998. Phylogenetic supertrees: Assembling the trees of life. Trends in Ecology & Evolution 13: p. 106. http: //www. cell. com/trends/ecology-evolution/abstract/S 0169 -5347%2897%2901242 -1 It is used subject to the fair use doctrine of: • Taiwan Copyright Act Articles 52 & 65 • Cell Press Copyrights and Limitations on Use • Elsevier Website Terms and Conditions • The "Code of Best Practices in Fair Use for Open. Course. Ware 2009 (http: //www. centerforsocialmedia. org/sites/default/files/10 -305 -OCW-Oct 29. pdf )" by A Committee of Practitioners of Open. Course. Ware in the U. S. The contents are based on Section 107 of the 1976 U. S. Copyright Act P 14 M. J. Sanderson, A. Purvis, and C. Henze 1998. Phylogenetic supertrees: Assembling the trees of life. Trends in Ecology & Evolution 13: p. 107. http: //www. cell. com/trends/ecology-evolution/abstract/S 0169 -5347%2897%2901242 -1 It is used subject to the fair use doctrine of: • Taiwan Copyright Act Articles 52 & 65 • Cell Press Copyrights and Limitations on Use • Elsevier Website Terms and Conditions • The "Code of Best Practices in Fair Use for Open. Course. Ware 2009 (http: //www. centerforsocialmedia. org/sites/default/files/10 -305 -OCW-Oct 29. pdf )" by A Committee of Practitioners of Open. Course. Ware in the U. S. The contents are based on Section 107 of the 1976 U. S. Copyright Act P 15 O. R. P. Bininda-Emonds, J. L. Gittleman, and M. A. Steel 2002. THE (SUPER)TREE OF LIFE: Procedures, Problems, and Prospects. Annual Review of Ecology and Systematics 33: p. 268. http: //www. annualreviews. org/doi/pdf/10. 1146/annurev. ecolsys. 33. 010802. 150511 It is used subject to the fair use doctrine of: • Taiwan Copyright Act Articles 52 & 65 • Annual Reviews Copyright & Permissions http: //www. annualreviews. org/page/about/copyright-and-permissions P 17 61

Work Licensing Author/Source Page P 18 National Taiwan University Jer-Ming Hu O. R. P. Bininda-Emonds, J. L. Gittleman, and M. A. Steel 2002. THE (SUPER)TREE OF LIFE: Procedures, Problems, and Prospects. Annual Review of Ecology and Systematics 33: p. 273. http: //www. annualreviews. org/doi/abs/10. 1146/annurev. ecolsys. 33. 010802. 150511? journal. Co de=ecolsys. 1 It is used subject to the fair use doctrine of: • Taiwan Copyright Act Articles 52 & 65 • Annual Reviews Copyright & Permissions http: //www. annualreviews. org/page/about/copyright-and-permissions P 19 O. R. P. Bininda-Emonds, J. L. Gittleman, and M. A. Steel 2002. THE (SUPER)TREE OF LIFE: Procedures, Problems, and Prospects. Annual Review of Ecology and Systematics 33: p. 273. http: //www. annualreviews. org/doi/abs/10. 1146/annurev. ecolsys. 33. 010802. 150511? journal. Co de=ecolsys. 1 It is used subject to the fair use doctrine of: • Taiwan Copyright Act Articles 52 & 65 • Annual Reviews Copyright & Permissions http: //www. annualreviews. org/page/about/copyright-and-permissions P 19 P 20 National Taiwan University Jer-Ming Hu O. R. P. Bininda-Emonds 2004. The evolution of supertrees. Trends in Ecology & Evolution 19: p. 317. http: //www. sciencedirect. com/science/article/pii/S 0169534704000709 It is used subject to the fair use doctrine of: • Taiwan Copyright Act Articles 52 & 65 • Elsevier Website Terms and Conditions http: //www. elsevier. com/wps/find/termsconditions. cws_home/termsconditions 62 P 21

Work Licensing Author/Source Page O. R. P. Bininda-Emonds 2004. The evolution of supertrees. Trends in Ecology & Evolution 19: p. 318. http: //www. sciencedirect. com/science/article/pii/S 0169534704000709 It is used subject to the fair use doctrine of: • Taiwan Copyright Act Articles 52 & 65 • Elsevier Website Terms and Conditions http: //www. elsevier. com/wps/find/termsconditions. cws_home/termsconditions P 22 O. R. P. Bininda-Emonds 2004. The evolution of supertrees. Trends in Ecology & Evolution 19: p. 320. http: //www. sciencedirect. com/science/article/pii/S 0169534704000709 It is used subject to the fair use doctrine of: • Taiwan Copyright Act Articles 52 & 65 • Elsevier Website Terms and Conditions http: //www. elsevier. com/wps/find/termsconditions. cws_home/termsconditions P 24 O. R. P. Bininda-Emonds 2004. The evolution of supertrees. Trends in Ecology & Evolution 19: p. 320. http: //www. sciencedirect. com/science/article/pii/S 0169534704000709 It is used subject to the fair use doctrine of: • Taiwan Copyright Act Articles 52 & 65 • Elsevier Website Terms and Conditions http: //www. elsevier. com/wps/find/termsconditions. cws_home/termsconditions P 25 M. J. Sanderson, A. Purvis, and C. Henze 1998. Phylogenetic supertrees: Assembling the trees of life. Trends in Ecology & Evolution 13: p. 108. http: //download. cell. com/trends/ecology-evolution/pdf/PIIS 0169534797012421. pdf It is used subject to the fair use doctrine of: • Taiwan Copyright Act Articles 52 & 65 • Cell Press Copyrights and Limitations on Use • Elsevier Website Terms and Conditions • The "Code of Best Practices in Fair Use for Open. Course. Ware 2009 (http: //www. centerforsocialmedia. org/sites/default/files/10 -305 -OCW-Oct 29. pdf )" by A Committee of Practitioners of Open. Course. Ware in the U. S. The contents are based on Section 107 of the 1976 U. S. Copyright Act P 29 63

Work Supertree using MRP 730 source trees, … Principal Coordinates analysis (PCO) for comparing tree distance (Robinson-Foulds distance) Licensing Author/Source Page M. J. Sanderson, A. Purvis, and C. Henze 1998. Phylogenetic supertrees: Assembling the trees of life. Trends in Ecology & Evolution 13: p. 108. http: //download. cell. com/trends/ecology-evolution/pdf/PIIS 0169534797012421. pdf It is used subject to the fair use doctrine of: • Taiwan Copyright Act Articles 52 & 65 • Cell Press Copyrights and Limitations on Use • Elsevier Website Terms and Conditions • The "Code of Best Practices in Fair Use for Open. Course. Ware 2009 (http: //www. centerforsocialmedia. org/sites/default/files/10 -305 -OCW-Oct 29. pdf )" by A Committee of Practitioners of Open. Course. Ware in the U. S. The contents are based on Section 107 of the 1976 U. S. Copyright Act P 29 Vincent Daubin, Manolo Gouy, and Guy Perriere A Phylogenomic Approach to Bacterial Phylogeny: Evidence of a Core of Genes Sharing a Common History http: //genome. cshlp. org/content/12/7/1080. full 2012/06/14 visited P 30 V. Daubin, M. Gouy, and G. Perriere. 2002. A Phylogenomic Approach to Bacterial Phylogeny: Evidence of a Core of Genes Sharing a Common History. Genome Research 12: p. 1082. Copyright © 2002, by Cold Spring Harbor Laboratory Press http: //genome. cshlp. org/content/12/7/1080. full. pdf+html 2012/06/14 visited P 31 V. Daubin, M. Gouy, and G. Perriere. 2002. A Phylogenomic Approach to Bacterial Phylogeny: Evidence of a Core of Genes Sharing a Common History. Genome Research 12: p. 1083. Copyright © 2002, by Cold Spring Harbor Laboratory Press http: //genome. cshlp. org/content/12/7/1080. full. pdf+html 2012/06/14 visited P 32 64

Work Supertree using MRP 46 source trees, include 379 taxa Bootstrap % used as character weights Examining diversification rates Licensing Author/Source Page V. Daubin, M. Gouy, and G. Perriere. 2002. A Phylogenomic Approach to Bacterial Phylogeny: Evidence of a Core of Genes Sharing a Common History. Genome Research 12: p. 1084. Copyright © 2002, by Cold Spring Harbor Laboratory Press http: //genome. cshlp. org/content/12/7/1080. full. pdf 2012/06/14 visited P 33 V. T. J. Davies, T. G. Barraclough, M. W. Chase, P. S. Soltis, D. E. Soltis, and V. Savolainen 2004. Darwin's abominable mystery: Insights from a supertree of the angiosperms. PNAS 101: p. 1904 -1909. http: //www. pnas. org/content/101/7/1904. full. pdf+html It is used subject to the fair use doctrine of: • Taiwan Copyright Act Articles 52 & 65 • The National Academies Terms of Use • The "Code of Best Practices in Fair Use for Open. Course. Ware 2009 (http: //www. centerforsocialmedia. org/sites/default/files/10 -305 -OCW-Oct 29. pdf )" by A Committee of Practitioners of Open. Course. Ware in the U. S. The contents are based on Section 107 of the 1976 U. S. Copyright Act P 34 V. T. J. Davies, T. G. Barraclough, M. W. Chase, P. S. Soltis, D. E. Soltis, and V. Savolainen 2004. Darwin's abominable mystery: Insights from a supertree of the angiosperms. PNAS 101: p. 1906. http: //www. pnas. org/content/101/7/1904. full. pdf+html It is used subject to the fair use doctrine of: • Taiwan Copyright Act Articles 52 & 65 • The National Academies Terms of Use • The "Code of Best Practices in Fair Use for Open. Course. Ware 2009 (http: //www. centerforsocialmedia. org/sites/default/files/10 -305 -OCW-Oct 29. pdf )" by A Committee of Practitioners of Open. Course. Ware in the U. S. The contents are based on Section 107 of the 1976 U. S. Copyright Act P 36 V. T. J. Davies, T. G. Barraclough, M. W. Chase, P. S. Soltis, D. E. Soltis, and V. Savolainen 2004. Darwin's abominable mystery: Insights from a supertree of the angiosperms. PNAS 101: p. 1908. http: //www. pnas. org/content/101/7/1904. full. pdf+html It is used subject to the fair use doctrine of: • Taiwan Copyright Act Articles 52 & 65 • The National Academies Terms of Use • The "Code of Best Practices in Fair Use for Open. Course. Ware 2009 (http: //www. centerforsocialmedia. org/sites/default/files/10 -305 -OCW-Oct 29. pdf )" by A Committee of Practitioners of Open. Course. Ware in the U. S. The contents are based on Section 107 of the 1976 U. S. Copyright Act P 37 P 39 National Taiwan University Jer-Ming Hu 65

Work Licensing Author/Source Page D. T. Pride, R. J. Meinersmann, T. M. Wassenaar, and M. J. Blaser 2003. Evolutionary Implications of Microbial Genome Tetranucleotide Frequency Biases. Genome Research 13: p. 152. http: //genome. cshlp. org/content/13/2/145. full. pdf+html 2012/06/14 visited P 41 D. T. Pride, R. J. Meinersmann, T. M. Wassenaar, and M. J. Blaser 2001. Genome trees constructed using five different approaches suggest new major bacterial clades. BMC Evolutionary Biology 1: 8. http: //www. biomedcentral. com/content/pdf/1471 -2148 -1 -8. pdf 2012/06/14 visited P 43 J. O. Korbel, B. Snel, M. A. Huynen, and P. Bork 2002. SHOT: a web server for the construction of genome phylogenies. Trends in Genetics 18: p. 160. http: //www. cell. com/trends/genetics/abstract/S 0168 -9525%2801%2902597 -5 It is used subject to the fair use doctrine of: • Taiwan Copyright Act Articles 52 & 65 • Cell Press Copyrights and Limitations on Use • Elsevier Website Terms and Conditions • The "Code of Best Practices in Fair Use for Open. Course. Ware 2009 (http: //www. centerforsocialmedia. org/sites/default/files/10 -305 -OCW-Oct 29. pdf )" by A Committee of Practitioners of Open. Course. Ware in the U. S. The contents are based on Section 107 of the 1976 U. S. Copyright Act P 43 J. O. Korbel, B. Snel, M. A. Huynen, and P. Bork 2002. SHOT: a web server for the construction of genome phylogenies. Trends in Genetics 18: p. 161. http: //www. cell. com/trends/genetics/abstract/S 0168 -9525%2801%2902597 -5 It is used subject to the fair use doctrine of: • Taiwan Copyright Act Articles 52 & 65 • Cell Press Copyrights and Limitations on Use • Elsevier Website Terms and Conditions • The "Code of Best Practices in Fair Use for Open. Course. Ware 2009 (http: //www. centerforsocialmedia. org/sites/default/files/10 -305 -OCW-Oct 29. pdf )" by A Committee of Practitioners of Open. Course. Ware in the U. S. The contents are based on Section 107 of the 1976 U. S. Copyright Act P 43, P 46 66

Work Licensing Author/Source Page B. Snel, P. Bork, M. A. Huynen 1999. Genome phylogeny based on gene content, Nature Genetics 21: p. 108. http: //www. nature. com/ng/journal/v 21/n 1/pdf/ng 0199_108. pdf It is used subject to the fair use doctrine of: • Taiwan Copyright Act Articles 52 & 65 • Nature Terms and Conditions P 45 J. O. Korbel, B. Snel, M. A. Huynen, and P. Bork 2002. SHOT: a web server for the construction of genome phylogenies. Trends in Genetics 18: p. 159. http: //www. cell. com/trends/genetics/pdf/PIIS 0168952501025975. pdf It is used subject to the fair use doctrine of: • Taiwan Copyright Act Articles 52 & 65 • Cell Press Copyrights and Limitations on Use • Elsevier Website Terms and Conditions • The "Code of Best Practices in Fair Use for Open. Course. Ware 2009 (http: //www. centerforsocialmedia. org/sites/default/files/10 -305 -OCW-Oct 29. pdf )" by A Committee of Practitioners of Open. Course. Ware in the U. S. The contents are based on Section 107 of the 1976 U. S. Copyright Act P 46 R. Delsuc, H. Brinkmann, and H. Philippe 2005. Phylogenomics and the reconstruction of the tree of life. Nature Reviews Genetics 6: p. 363. http: //www. nature. com/nrg/journal/v 6/n 5/pdf/nrg 1603. pdf It is used subject to the fair use doctrine of: • Taiwan Copyright Act Articles 52 & 65 • Nature Terms and Conditions P 49 G. D. Edgecombe, G. Giribet, C. W. Dunn, A. Hejnol, R. M. Kristensen, R. C. Neves, G. W. Rouse, K. Worsaae, and M. V. Sørensen Organisms Diversity & Evolution 11: p. 155. http: //www. springerlink. com/content/5608113750 q 30314/fulltext. pdf? MUD=MP This work is licensed by Springer. Link for the use of “Course Database of General Education TW” ONLY. The copyright belongs to the above mentioned entity and GET does not have the authorization right. Copyright privileges have to be negotiated with the copyright owner(s) for separately. P 50 67

Work Licensing Author/Source Page G. D. Edgecombe, G. Giribet, C. W. Dunn, A. Hejnol, R. M. Kristensen, R. C. Neves, G. W. Rouse, K. Worsaae, and M. V. Sørensen Organisms Diversity & Evolution 11: p. 155. http: //www. springerlink. com/content/5608113750 q 30314/fulltext. pdf? MUD=MP This work is licensed by Springer. Link for the use of “Course Database of General Education TW” ONLY. The copyright belongs to the above mentioned entity and GET does not have the authorization right. Copyright privileges have to be negotiated with the copyright owner(s) for separately. P 50 C. W. Dunn, A. Hejnol, D. Q. Matus, K. Pang, W. E. Browne, S. A. Smith, E. Seaver, G. W. Rouse, M. Obst, G. D. Edgecombe, M. V. Sørensen 6, S. H. D. Haddock, A. Schmidt-Rhaesa, A. Okusu, R. M. Kristensen, W. C. Wheeler, M. Q. Martindale, and G. Giribet 2008. Broad phylogenomic sampling improves resolution of the animal tree of life. Nature 452: p. 745 -349 http: //www. nature. com/nature/journal/v 452/n 7188/extref/nature 06614 -s 1. pdf It is used subject to the fair use doctrine of: • Taiwan Copyright Act Articles 52 & 65 • Nature Terms and Conditions P 51 C. W. Dunn, A. Hejnol, D. Q. Matus, K. Pang, W. E. Browne, S. A. Smith, E. Seaver, G. W. Rouse, M. Obst, G. D. Edgecombe, M. V. Sørensen 6, S. H. D. Haddock, A. Schmidt-Rhaesa, A. Okusu, R. M. Kristensen, W. C. Wheeler, M. Q. Martindale, and G. Giribet 2008. Broad phylogenomic sampling improves resolution of the animal tree of life. Nature 452: p. 746. http: //www. nature. com/nature/journal/v 452/n 7188/full/nature 06614. html? free=2 It is used subject to the fair use doctrine of: • Taiwan Copyright Act Articles 52 & 65 • Nature Terms and Conditions P 52 Bastien Boussau, and Vincent Daubin 2010. Genomes as documents of evolutionary history. Trends in Ecology & Evolution 25: p. 228. http: //www. sciencedirect. com/science/article/pii/S 0169534709002973 It is used subject to the fair use doctrine of: • Taiwan Copyright Act Articles 52 & 65 • Elsevier Website Terms and Conditions http: //www. elsevier. com/wps/find/termsconditions. cws_home/termsconditions P 53 68

Work Licensing Author/Source Page Bastien Boussau, and Vincent Daubin 2010. Genomes as documents of evolutionary history. Trends in Ecology & Evolution 25: p. 225. http: //www. sciencedirect. com/science/article/pii/S 0169534709002973 It is used subject to the fair use doctrine of: • Taiwan Copyright Act Articles 52 & 65 • Elsevier Website Terms and Conditions http: //www. elsevier. com/wps/find/termsconditions. cws_home/termsconditions P 54 P 55 Flickr scott. zona http: //www. flickr. com/photos/scottzona/3173820625/sizes/l/in/photostream/ 2012/06/14 visited P 55 Flickr adrien 2008 http: //www. flickr. com/photos/adrien 2008/4647588299/sizes/l/in/photostream/ 2012/06/14 visited P 56 S. Stefanović, D. W. Rice, and J. D. Palmer 2004. Long branch attraction, taxon sampling, and the earliest angiosperms: Amborella or monocots? BMC Evolutionary Biology 4: 35. http: //w 01. biomedcentral. com/content/pdf/1471 -2148 -4 -35. pdf 2012/06/14 visited P 56 Flickr pennstatelive http: //www. flickr. com/photos/pennstatelive/5609235215/sizes/l/in/photostream/ 2012/06/14 visited 69

Work Licensing Author/Source Page V. V. Goremykin, K. I. Hirsch-Ernst, S. Wolfl, and F. H. Hellwig 2004. The Chloroplast Genome of Nymphaea alba: Whole-Genome Analyses and the Problem of Identifying the Most Basal Angiosperm. Molecular Biology and Evolution 21: p. 1448. http: //mbe. oxfordjournals. org/content/21/7/1445. full It is used subject to the fair use doctrine of: • Taiwan Copyright Act Articles 52 & 65 • Oxford Journals Terms & Conditions for Use http: //www. oxfordjournals. org/our_journals/terms. html P 57 P 58 S. Stefanović, D. W. Rice, and J. D. Palmer 2004. Long branch attraction, taxon sampling, and the earliest angiosperms: Amborella or monocots? BMC Evolutionary Biology 4: 35. http: //w 01. biomedcentral. com/1471 -2148/4/35/ 2012/06/14 visited P 59 S. Stefanović, D. W. Rice, and J. D. Palmer 2004. Long branch attraction, taxon sampling, and the earliest angiosperms: Amborella or monocots? BMC Evolutionary Biology 4: 35. http: //w 01. biomedcentral. com/1471 -2148/4/35/ 2012/06/14 visited 70