Multiple Sequence Alignment CS 273 a Lecture 10


- Slides: 14

Multiple Sequence Alignment CS 273 a Lecture 10, Aut 08, Batzoglou

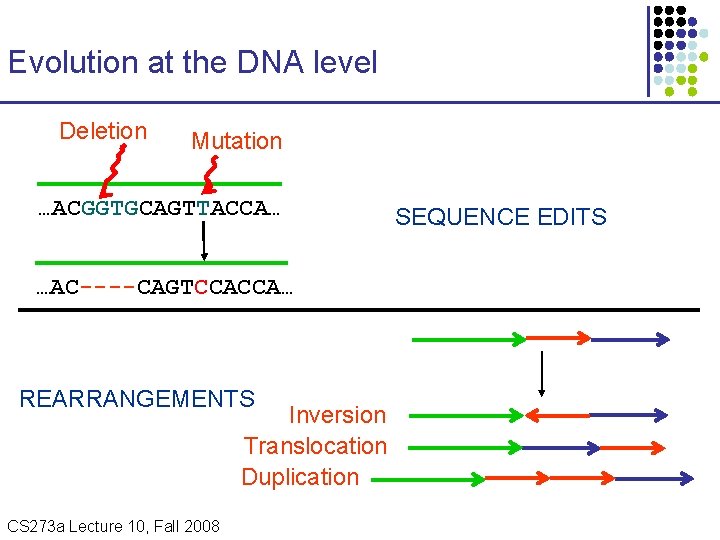
Evolution at the DNA level Deletion Mutation …ACGGTGCAGTTACCA… …AC----CAGTCCACCA… REARRANGEMENTS Inversion Translocation Duplication CS 273 a Lecture Fall 2008 CS 273 a Lecture 10, Aut 08, 10, Batzoglou SEQUENCE EDITS

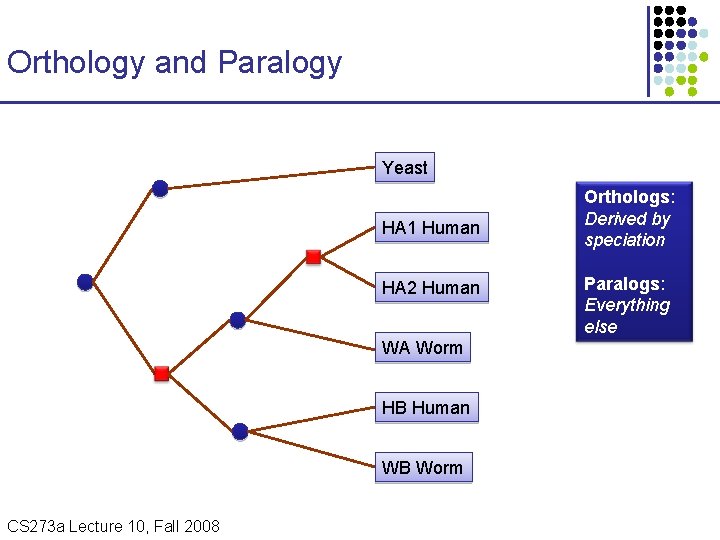
Orthology and Paralogy Yeast HA 1 Human HA 2 Human WA Worm HB Human WB Worm CS 273 a Lecture Fall 2008 CS 273 a Lecture 10, Aut 08, 10, Batzoglou Orthologs: Derived by speciation Paralogs: Everything else

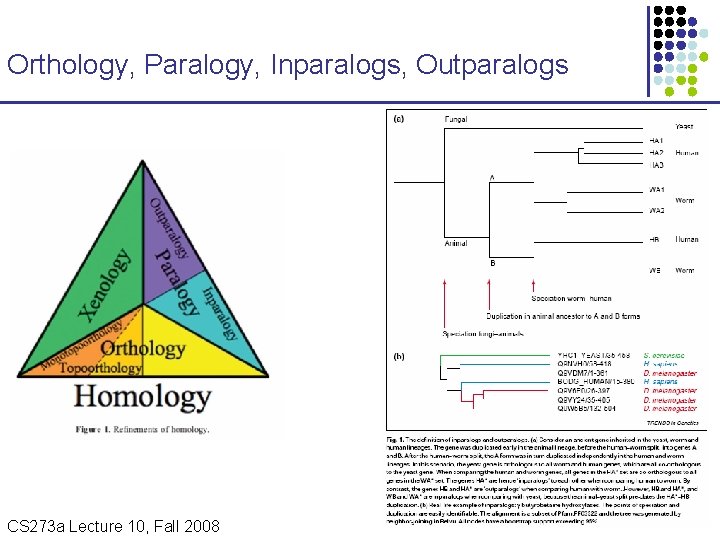
Orthology, Paralogy, Inparalogs, Outparalogs CS 273 a Lecture Fall 2008 CS 273 a Lecture 10, Aut 08, 10, Batzoglou

CS 273 a Lecture Fall 2008 CS 273 a Lecture 10, Aut 08, 10, Batzoglou

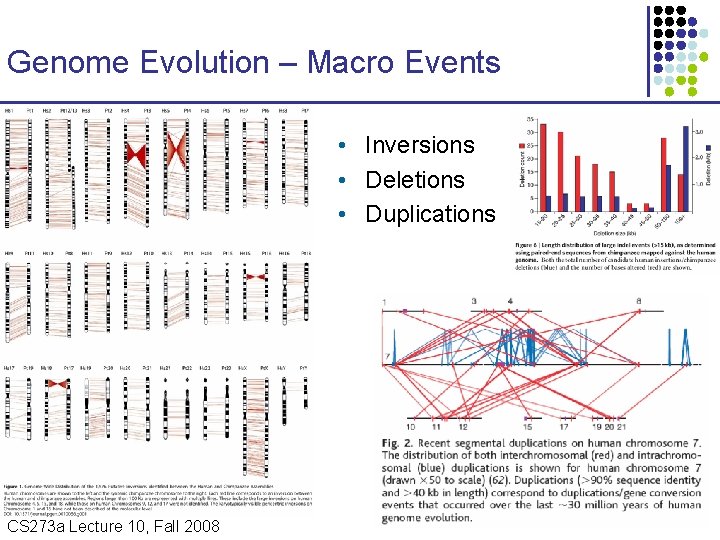
Genome Evolution – Macro Events • Inversions • Deletions • Duplications CS 273 a Lecture Fall 2008 CS 273 a Lecture 10, Aut 08, 10, Batzoglou

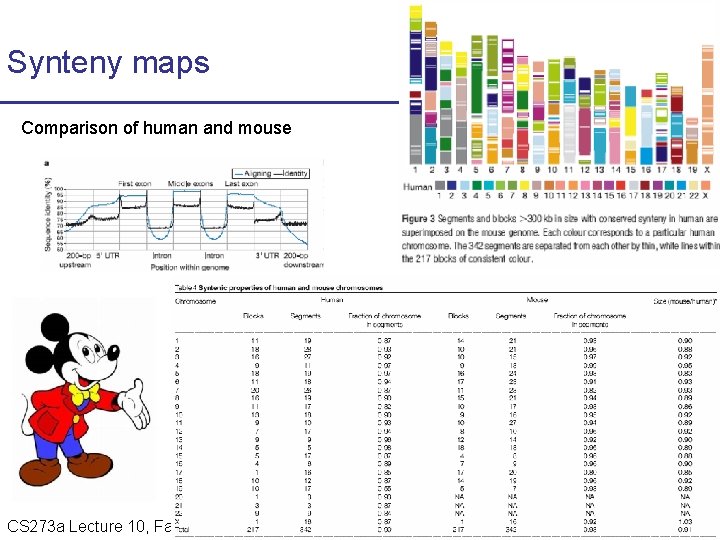
Synteny maps Comparison of human and mouse CS 273 a Lecture Fall 2008 CS 273 a Lecture 10, Aut 08, 10, Batzoglou

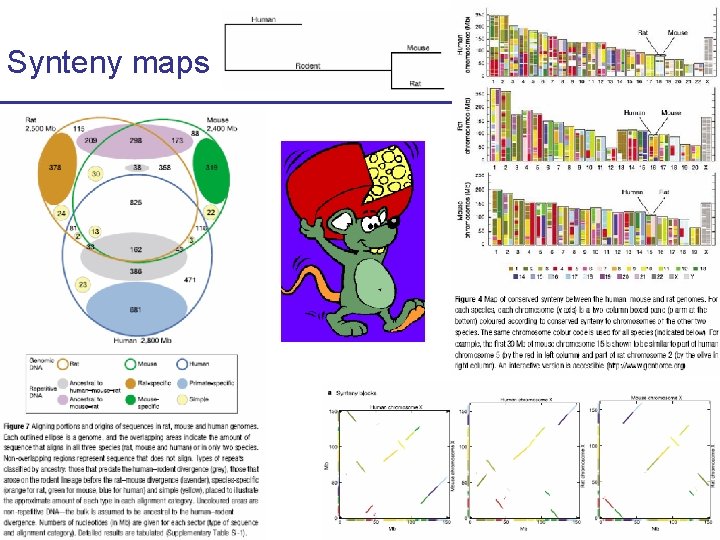
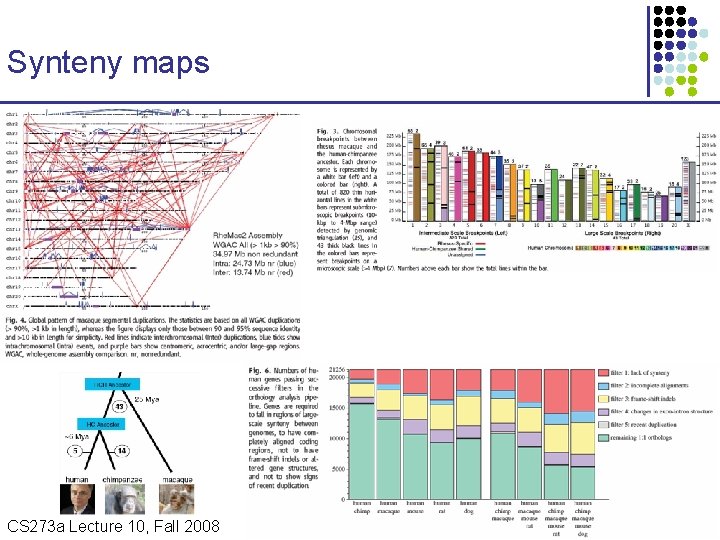
Synteny maps CS 273 a Lecture Fall 2008 CS 273 a Lecture 10, Aut 08, 10, Batzoglou

Synteny maps CS 273 a Lecture Fall 2008 CS 273 a Lecture 10, Aut 08, 10, Batzoglou

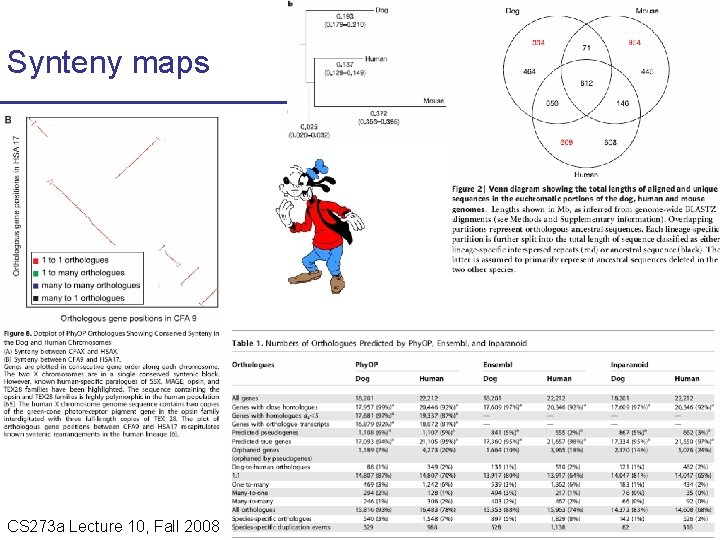
Synteny maps CS 273 a Lecture Fall 2008 CS 273 a Lecture 10, Aut 08, 10, Batzoglou

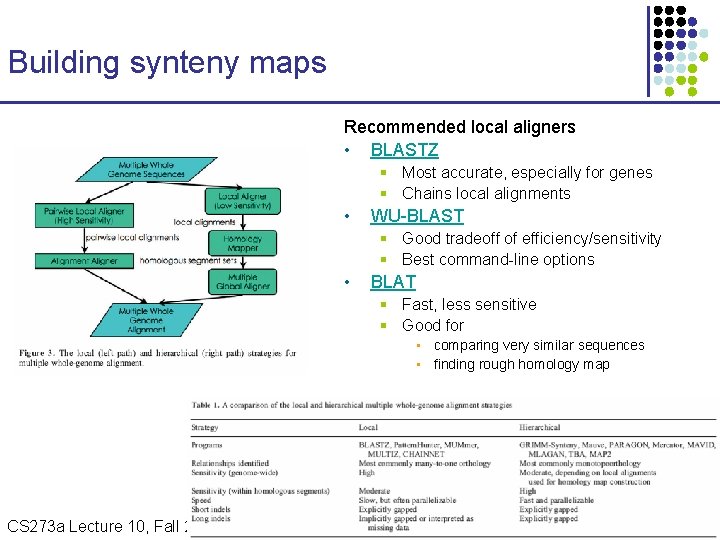
Building synteny maps Recommended local aligners • BLASTZ § Most accurate, especially for genes § Chains local alignments • WU-BLAST § Good tradeoff of efficiency/sensitivity § Best command-line options • BLAT § Fast, less sensitive § Good for • comparing very similar sequences • finding rough homology map CS 273 a Lecture Fall 2008 CS 273 a Lecture 10, Aut 08, 10, Batzoglou

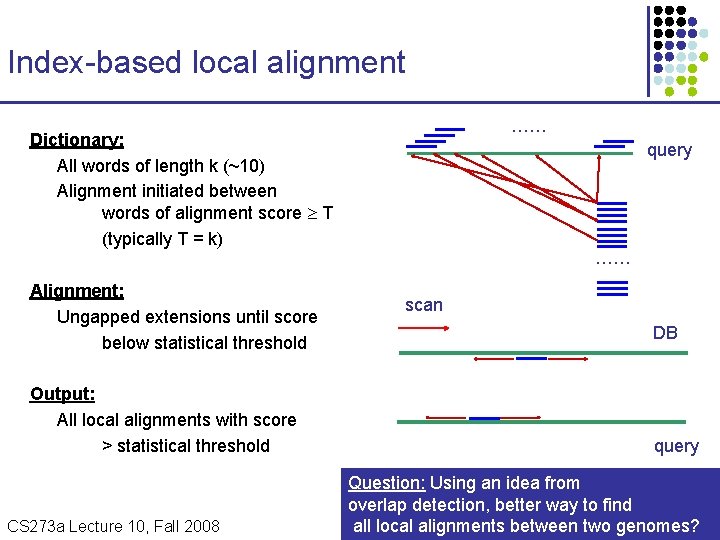
Index-based local alignment …… Dictionary: All words of length k (~10) Alignment initiated between words of alignment score T (typically T = k) Alignment: Ungapped extensions until score below statistical threshold Output: All local alignments with score > statistical threshold CS 273 a Lecture Fall 2008 CS 273 a Lecture 10, Aut 08, 10, Batzoglou query …… scan DB query Question: Using an idea from overlap detection, better way to find all local alignments between two genomes?

Local Alignments CS 273 a Lecture Fall 2008 CS 273 a Lecture 10, Aut 08, 10, Batzoglou

After chaining CS 273 a Lecture Fall 2008 CS 273 a Lecture 10, Aut 08, 10, Batzoglou