Microarray Data Analysis n n Data preprocessing and






- Slides: 24

Microarray Data Analysis n n Data preprocessing and visualization Supervised learning n n Unsupervised learning n n Machine learning approaches Clustering and pattern detection Gene regulatory regions predictions based coregulated genes Linkage between gene expression data and gene sequence/function databases …

Unsupervised learning n n Supervised methods n Can only validate or reject hypotheses n Can not lead to discovery of unexpected partitions Unsupervised learning n No prior knowledge is used n Explore structure of data on the basis of corrections and similarities

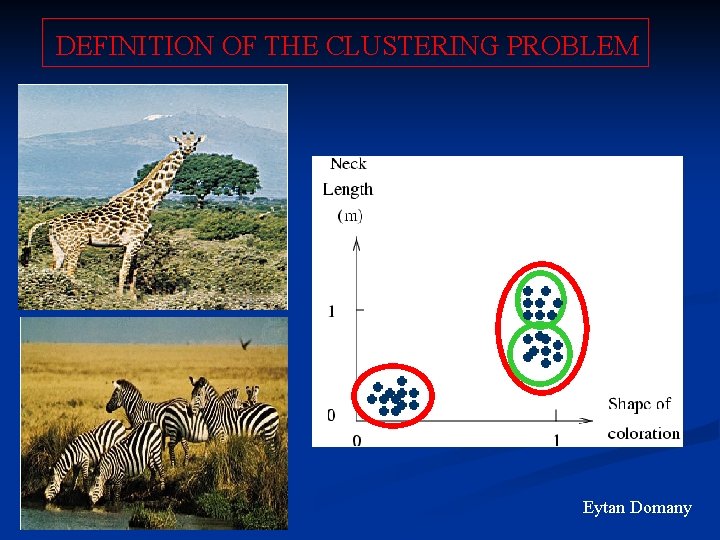
DEFINITION OF THE CLUSTERING PROBLEM Eytan Domany

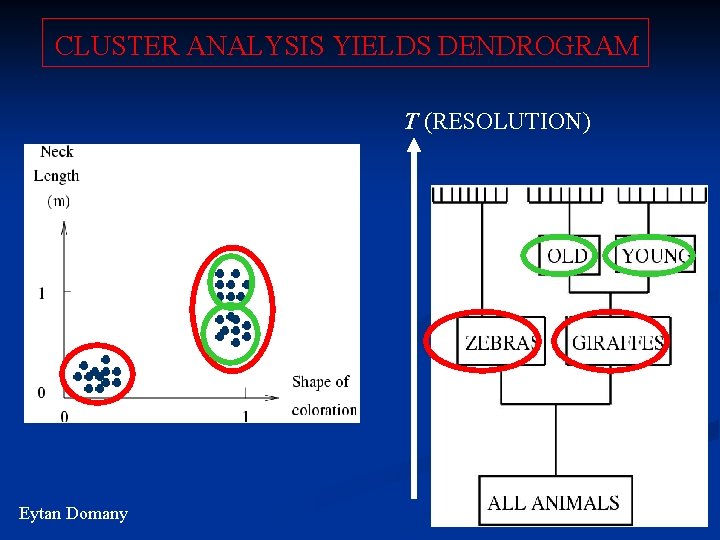
CLUSTER ANALYSIS YIELDS DENDROGRAM T (RESOLUTION) Eytan Domany

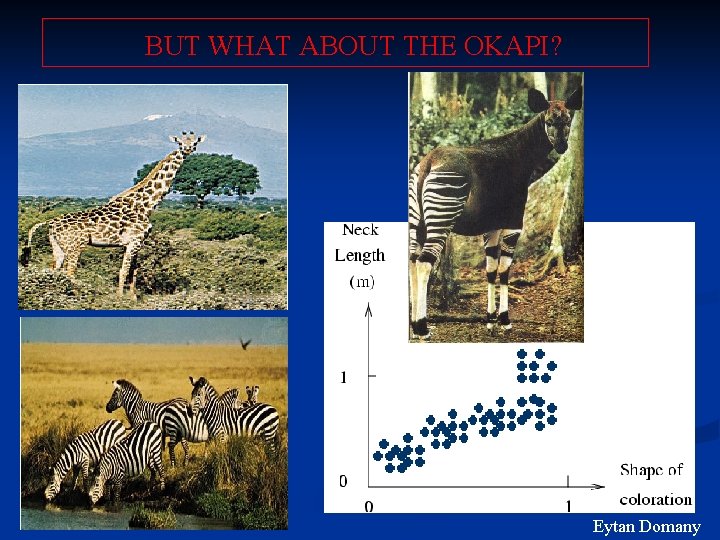
BUT WHAT ABOUT THE OKAPI? Eytan Domany

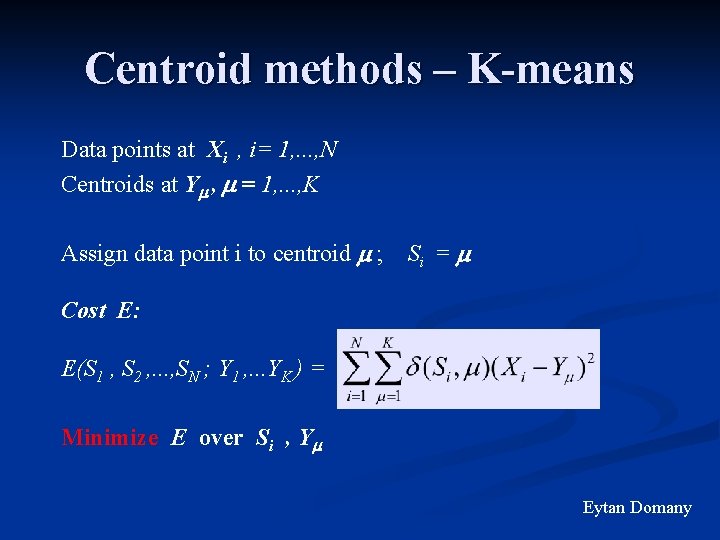
Centroid methods – K-means Data points at Xi , i= 1, . . . , N Centroids at Y , = 1, . . . , K Assign data point i to centroid ; Si = Cost E: E(S 1 , S 2 , . . . , SN ; Y 1 , . . . YK ) = Minimize E over Si , Y Eytan Domany

K-means n “Guess” K=3 Eytan Domany

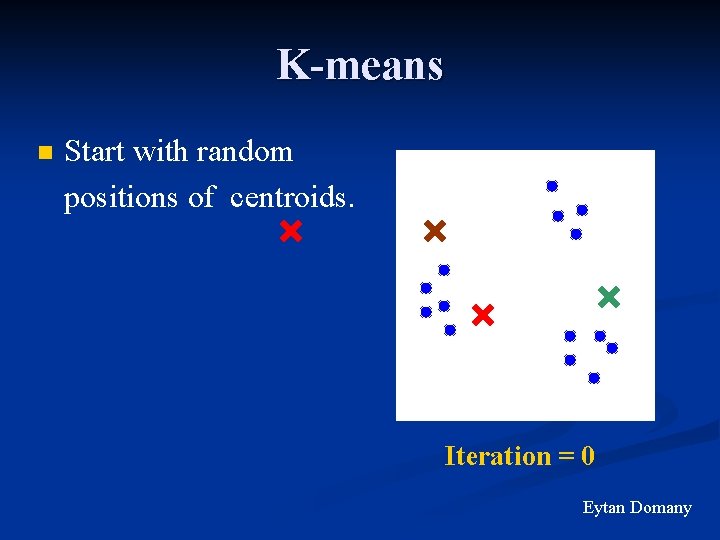
K-means n Start with random positions of centroids. Iteration = 0 Eytan Domany

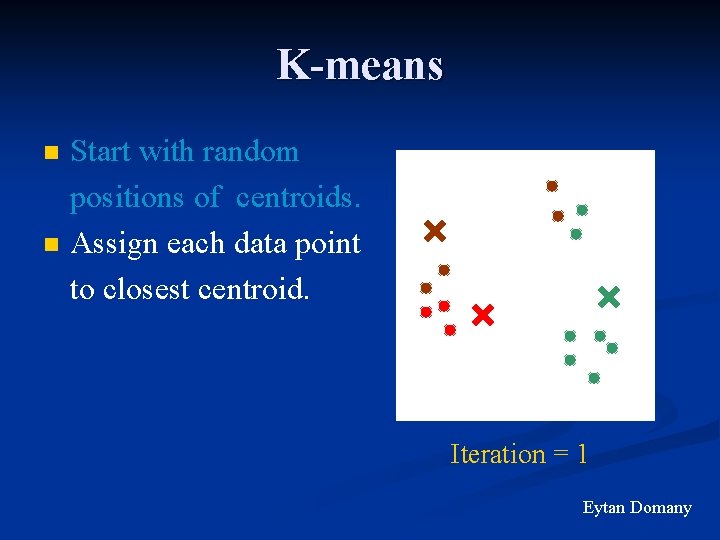
K-means n n Start with random positions of centroids. Assign each data point to closest centroid. Iteration = 1 Eytan Domany

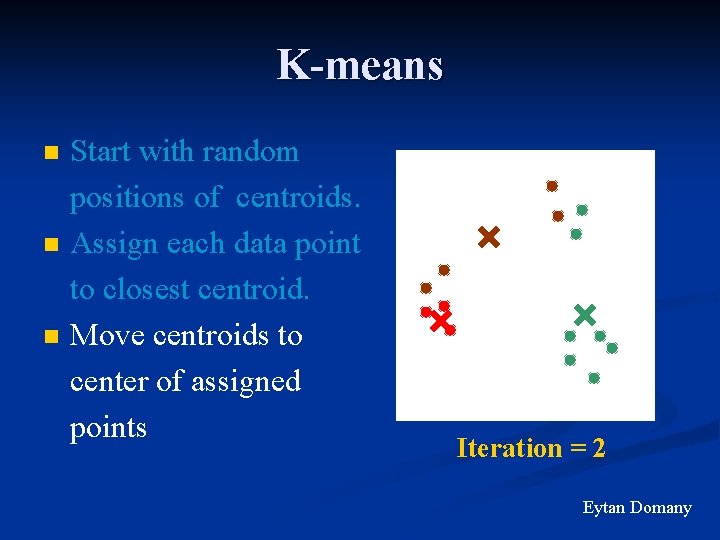
K-means n n n Start with random positions of centroids. Assign each data point to closest centroid. Move centroids to center of assigned points Iteration = 2 Eytan Domany

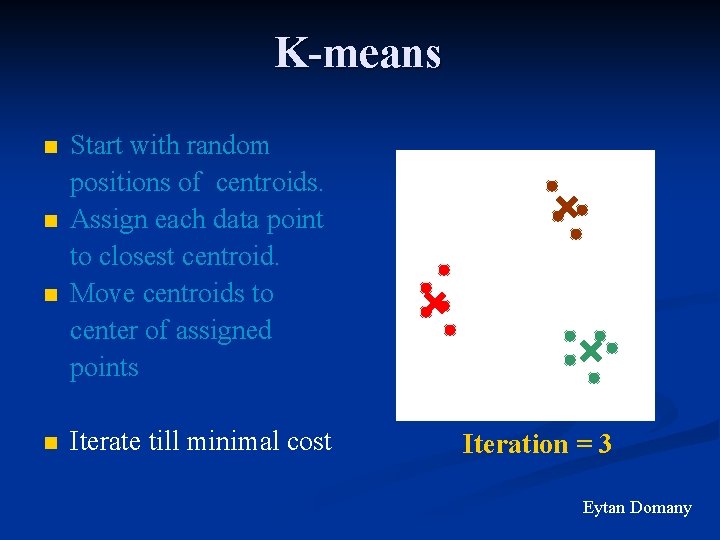
K-means n n Start with random positions of centroids. Assign each data point to closest centroid. Move centroids to center of assigned points Iterate till minimal cost Iteration = 3 Eytan Domany

K-means - Summary n Fast algorithm: compute distances from data points to centroids Result depends on initial centroids’ position n Must preset K n Fails for “non-spherical” distributions n

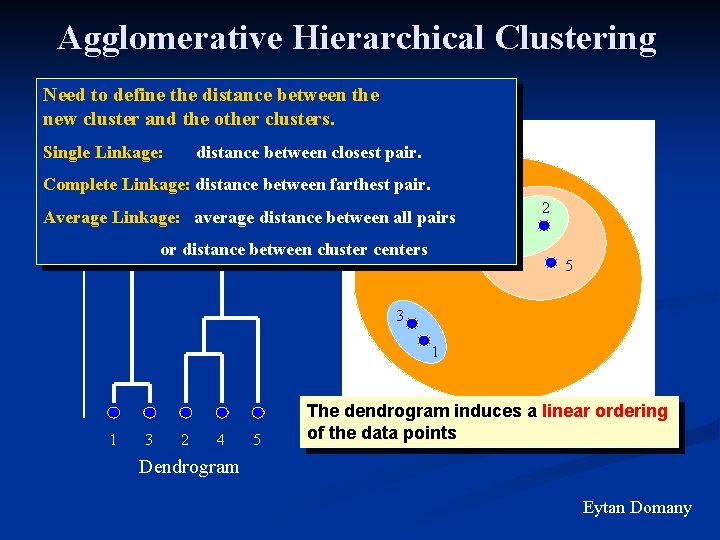
Agglomerative Hierarchical Clustering Need to define the distance between theclusters at each step merge pair of nearest new cluster and the other clusters. initially – each point = cluster Single Linkage: distance between closest pair. Distance between joined clusters Complete Linkage: distance between farthest pair. Average Linkage: average distance between all pairs or distance between cluster centers 4 2 5 3 1 1 3 2 4 5 The dendrogram induces a linear ordering of the data points Dendrogram Eytan Domany

Hierarchical Clustering Summary n Results depend on distance update method n Greedy iterative process n NOT robust against noise n No inherent measure to identify stable clusters n Average Linkage – the most widely used clustering method in gene expression analysis

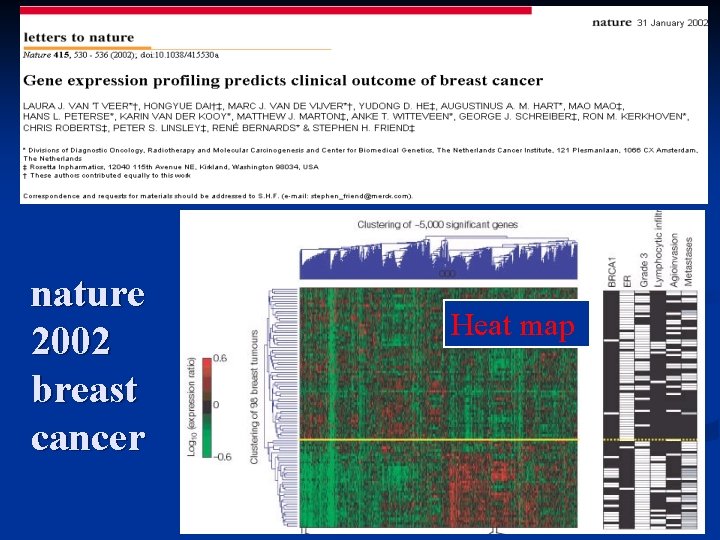
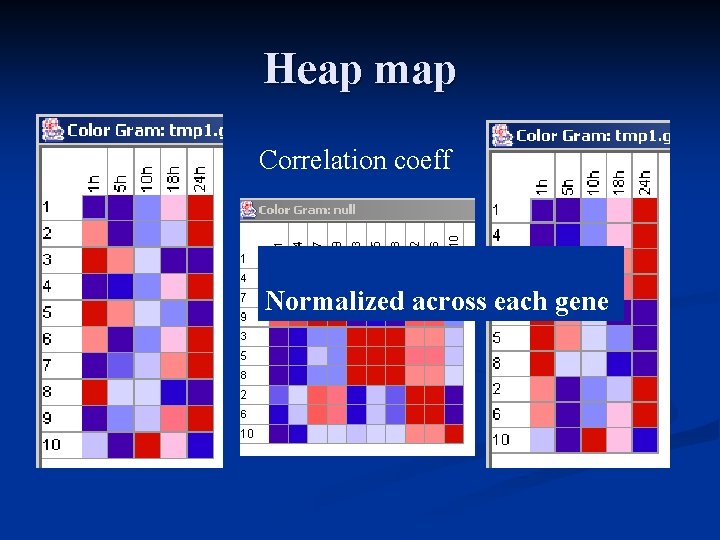
nature 2002 breast cancer Heat map

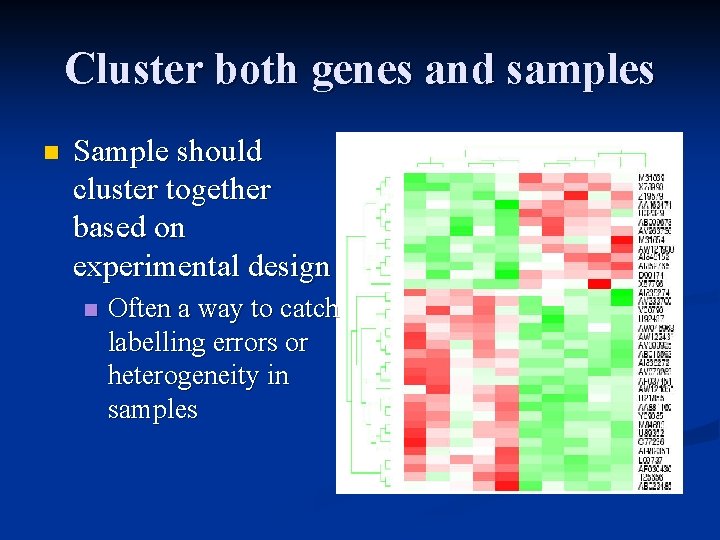
Cluster both genes and samples n Sample should cluster together based on experimental design n Often a way to catch labelling errors or heterogeneity in samples

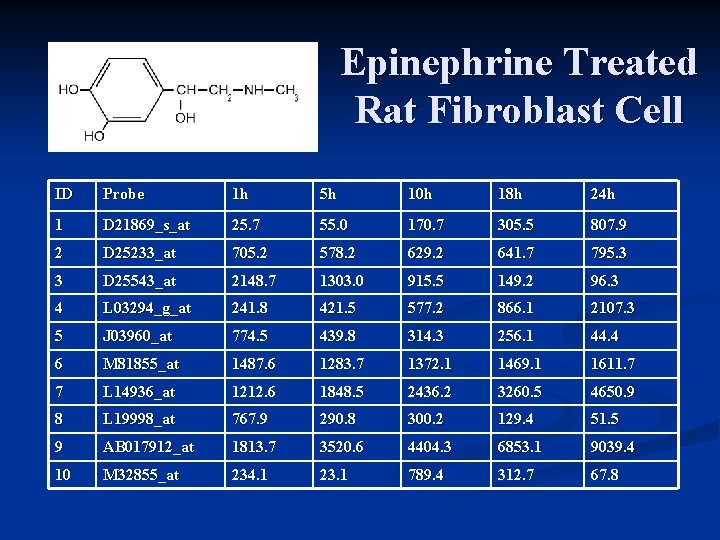
Epinephrine Treated Rat Fibroblast Cell ID Probe 1 h 5 h 10 h 18 h 24 h 1 D 21869_s_at 25. 7 55. 0 170. 7 305. 5 807. 9 2 D 25233_at 705. 2 578. 2 629. 2 641. 7 795. 3 3 D 25543_at 2148. 7 1303. 0 915. 5 149. 2 96. 3 4 L 03294_g_at 241. 8 421. 5 577. 2 866. 1 2107. 3 5 J 03960_at 774. 5 439. 8 314. 3 256. 1 44. 4 6 M 81855_at 1487. 6 1283. 7 1372. 1 1469. 1 1611. 7 7 L 14936_at 1212. 6 1848. 5 2436. 2 3260. 5 4650. 9 8 L 19998_at 767. 9 290. 8 300. 2 129. 4 51. 5 9 AB 017912_at 1813. 7 3520. 6 4404. 3 6853. 1 9039. 4 10 M 32855_at 234. 1 23. 1 789. 4 312. 7 67. 8

Heap map Correlation coeff Normalized across each gene

Distance Issues n Euclidean distance g 1 g 3 g 2 g 4 ■ Pearson distance

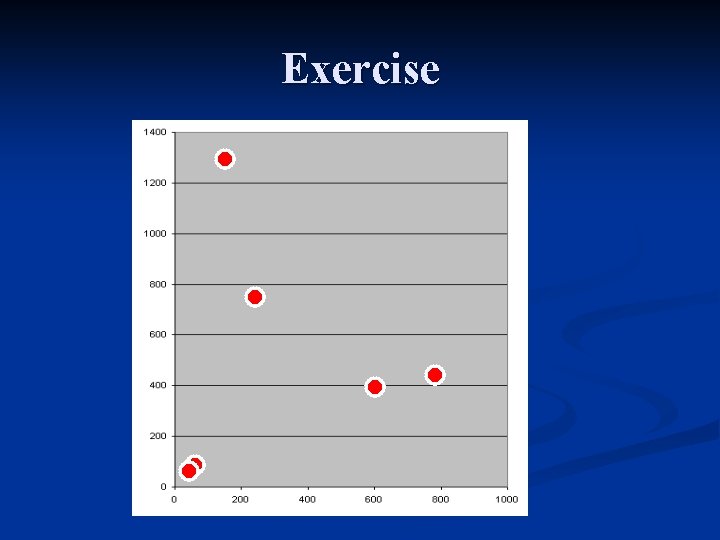
Exercise n Use Average Linkage Algorithm and Manhattan distance. Gene ID 1 2 3 Exp 1 45 55 148 Exp 2 55 78 1303 4 5 6 241 774 607 765 439 383

Exercise

Issues in Cluster Analysis A lot of clustering algorithms n A lot of distance/similarity metrics n Which clustering algorithm runs faster and uses less memory? n How many clusters after all? n Are the clusters stable? n Are the clusters meaningful? n

Which Clustering Method Should I Use? What is the biological question? n Do I have a preconceived notion of how many clusters there should be? n How strict do I want to be? Spilt or Join? n Can a gene be in multiple clusters? n Hard or soft boundaries between clusters n

The End n Thank you for taking this course. Bioinformatics is a very diverse and fascinating subject. We hope you all decide to continue your pursuit of it. n We will be very glad to answer your emails or schedule appointments to talk about any bioinformatics related questions you might have. n We wish you all have a wonderful summer break!