LECTURE 1 Phylogeny and Systematics What is Phylogeny








- Slides: 39

LECTURE 1: Phylogeny and Systematics

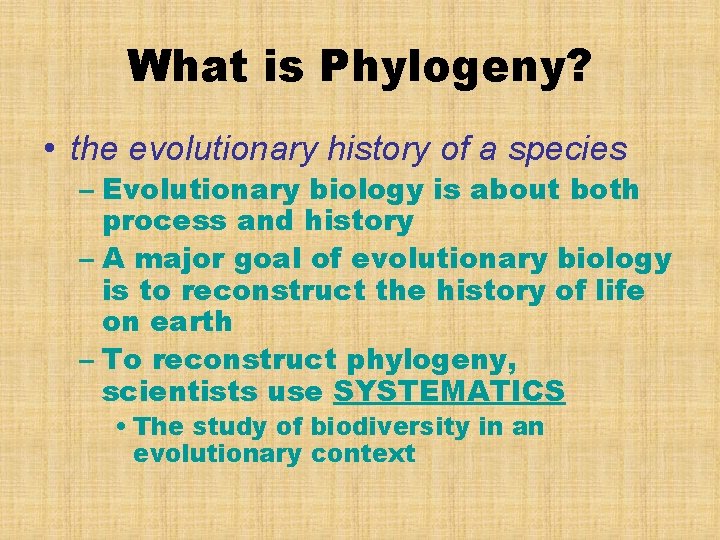
What is Phylogeny? • the evolutionary history of a species – Evolutionary biology is about both process and history – A major goal of evolutionary biology is to reconstruct the history of life on earth – To reconstruct phylogeny, scientists use SYSTEMATICS • The study of biodiversity in an evolutionary context

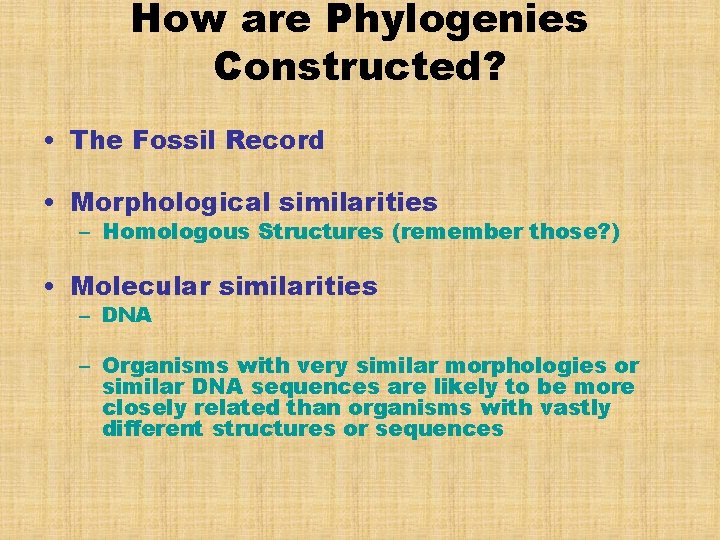
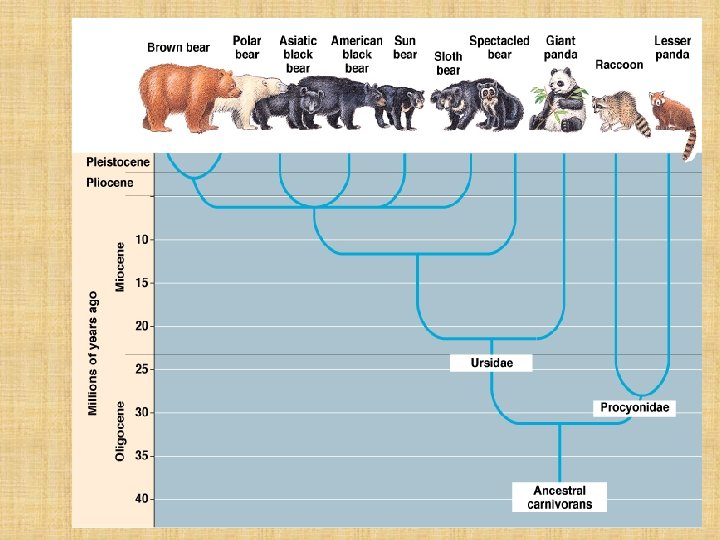
How are Phylogenies Constructed? • The Fossil Record • Morphological similarities – Homologous Structures (remember those? ) • Molecular similarities – DNA – Organisms with very similar morphologies or similar DNA sequences are likely to be more closely related than organisms with vastly different structures or sequences

How can Scientists determine whether structure are Homologous or Analagous? • HOMOLOGY is similarity due to SHARED ANCESTRY • ANALOGY is similarity due to CONVERGENT EVOLUTION – CONVERGENT EVOLUTION: two organisms develop similarities as they adapted to similar environmental challenges – not because they evolved from a common ancestor – EXAMPLE: both birds and bats have adaptations that allow them to fly – However, a close examination of a bat’s wing shows a greater similarity to a cat’s forelimb than to a bird’s wing – Fossil evidence also documents that bat and bird wings arose independently from walking forelimbs of different ancestors – Thus a bat’s wing is homologous to other mammalian forelimbs but is analogous in function to a bird’s wing

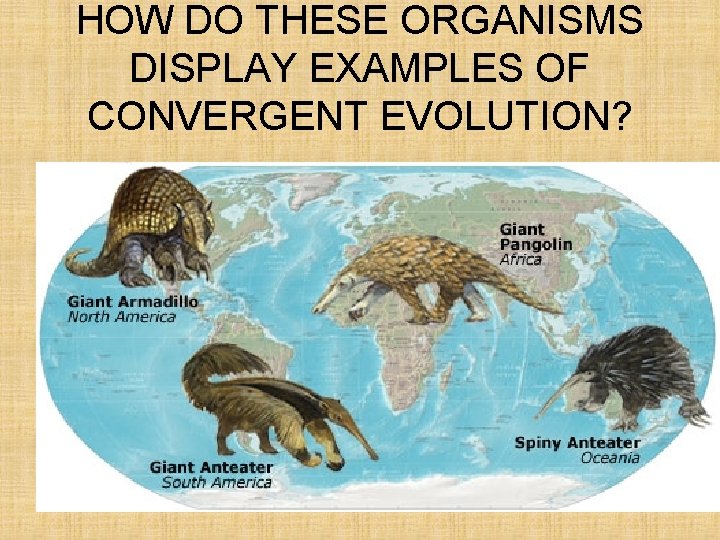
HOW DO THESE ORGANISMS DISPLAY EXAMPLES OF CONVERGENT EVOLUTION?

How would you compare the fins in these 2 organisms?

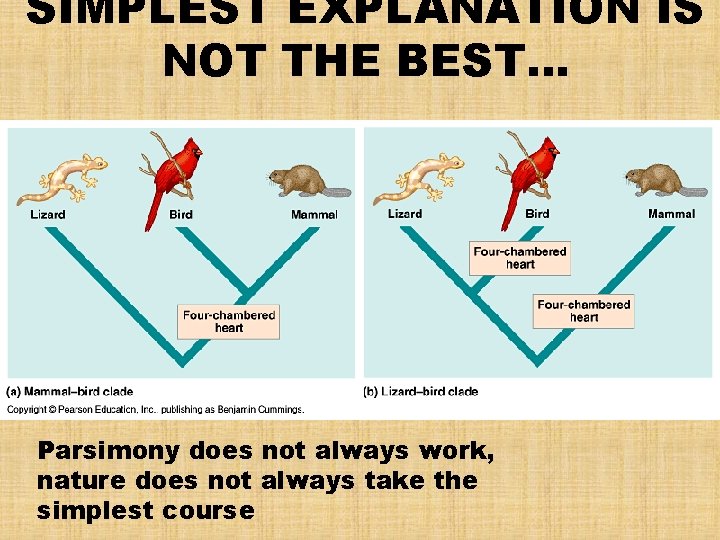
What are Homoplasies? • Analogous structures or molecular sequences that evolved independently • Example: the four-chambered heart of birds & mammals is analogous

What are Molecular Homologies? • Systematists compare long stretches of DNA and even entire genomes to assess relationships between species • If genes in two organisms have closely similar nucleotide sequences, it is highly likely that the genes are homologous • In closely related species, sequences may differ at only one or a few base sites • Distantly related species may have many differences or sequences of different length

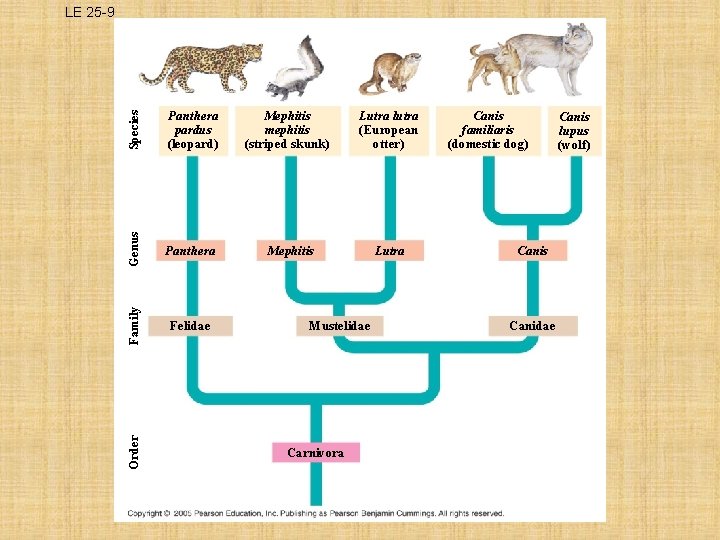
How is Phylogeny linked with Classification? Ø Systematists explore phylogeny by examining various characteristics in living and fossil organisms Ø They construct branching diagrams called PHYLOGENETIC TREES to depict their hypotheses about evolutionary relationships Ø The branching of the tree reflects the hierarchical classification of groups nested within more inclusive groups

Species Mephitis mephitis (striped skunk) Lutra lutra (European otter) Genus Panthera Mephitis Lutra Felidae Order Panthera pardus (leopard) Family LE 25 -9 Mustelidae Carnivora Canis familiaris (domestic dog) Canis Canidae Canis lupus (wolf)

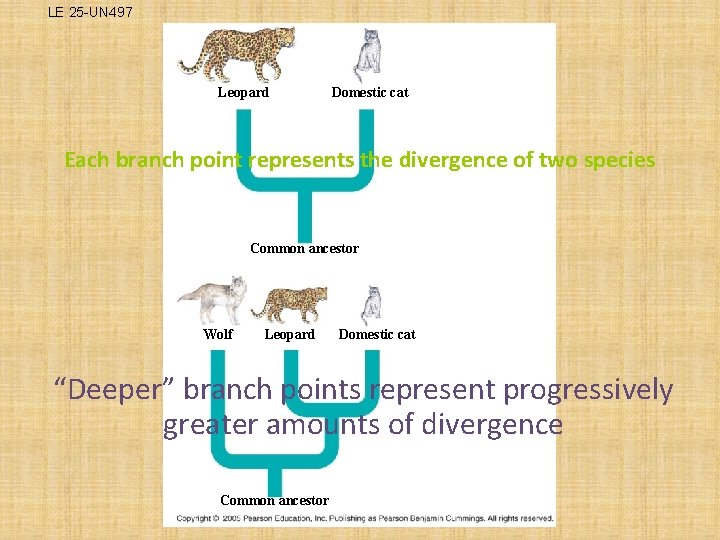
LE 25 -UN 497 Leopard Domestic cat Each branch point represents the divergence of two species Common ancestor Wolf Leopard Domestic cat “Deeper” branch points represent progressively greater amounts of divergence Common ancestor

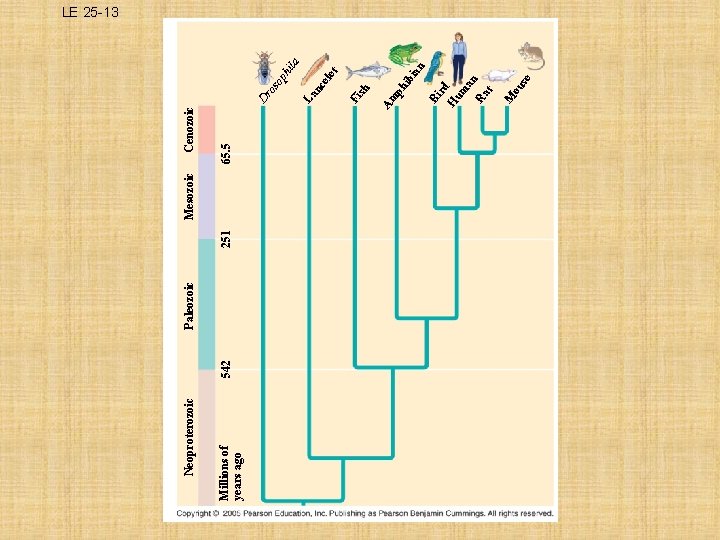
Millions of years ago Neoproterozoic 542 Paleozoic 251 Mesozoic 65. 5 bi an e ou s M Bi rd H um an Ra t ph i Fi sh let ce La n la op hi os Dr Am Cenozoic LE 25 -13

What is Cladistics? Ø classifying organisms based on RESEMBLANCES among clades § Determining which similarities between species are relevant to grouping the species in a clade is a challenge § It is especially important to distinguish similarities that are based on shared ancestry or homology from those that are based on convergent evolution or analogy CLADISTICS enables us to identify the sequence of the evolution of derived characteristics

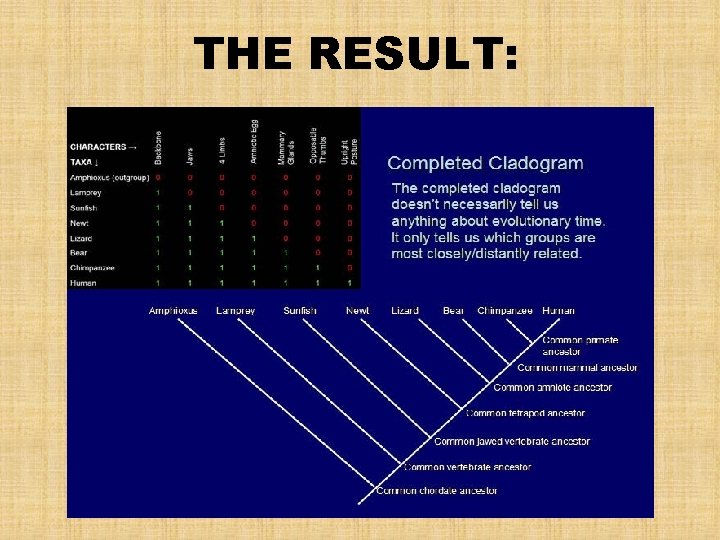
What is a Cladogram? • depicts patterns of shared derived characteristics among taxa – Synapomorphies • the chronological sequence of branching during the evolutionary history of a set of organisms • This chronology DOES NOT indicate the TIME of origin of the species that we are comparing, only the groups to which they belong • A cladogram is NOT a phylogenetic tree • To convert it to a phylogenetic tree, we need more information from sources such as the fossil record, which can indicate when and in which groups the characters first appeared

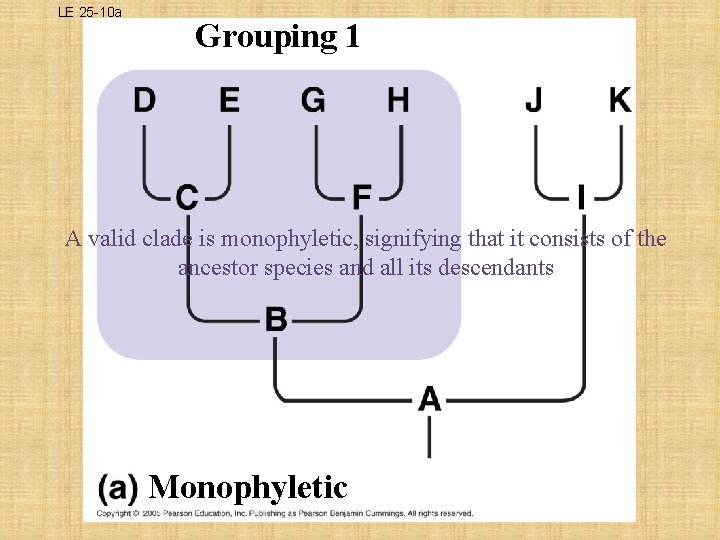
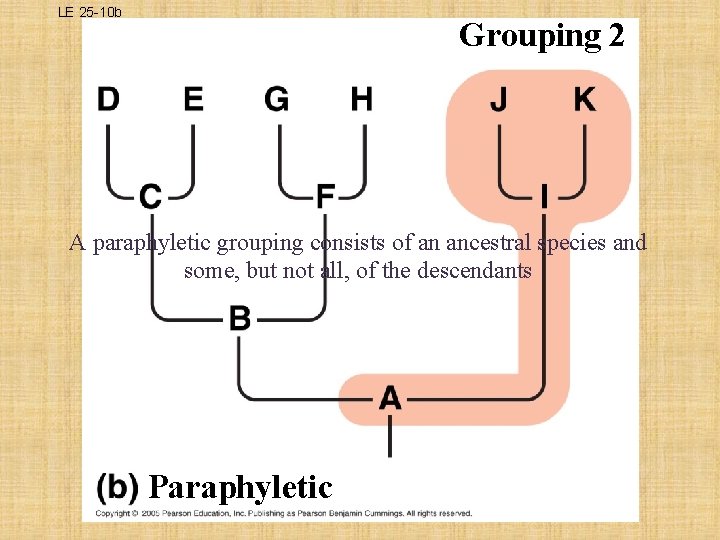
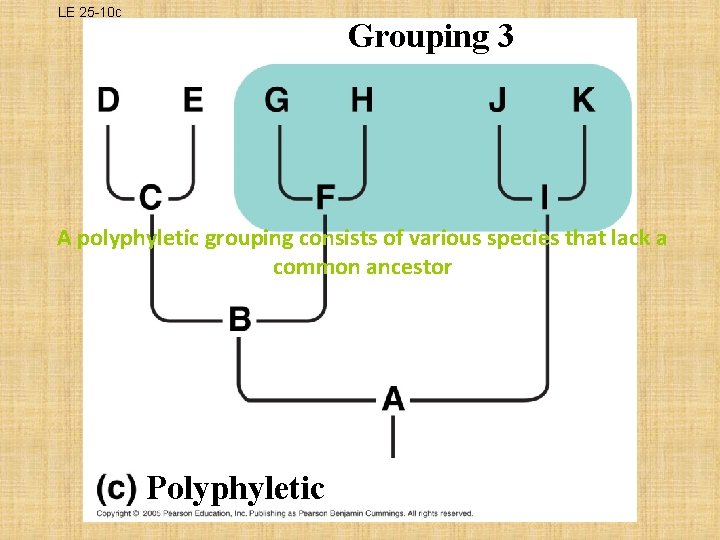
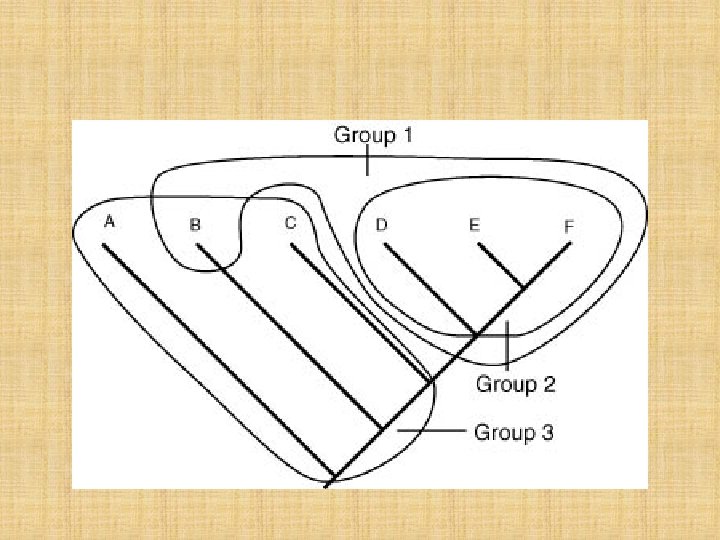
What is a Clade? Ø a group of species that includes an ancestral species and all its descendants • THREE TYPES OF CLADES: – MONOPHYLETIC • single ancestor that gives rise to all species in that taxon and to no species in any other taxon; legitimate cladogram – PARAPHYLETIC • members of a taxa are derived from 2 or more ancestral forms not common to all members; does not meet cladistic criterion – POLYPHYLETIC • lacks the common ancestor that would unite the species; does not meet cladistic criterion

LE 25 -10 a Grouping 1 A valid clade is monophyletic, signifying that it consists of the ancestor species and all its descendants Monophyletic

LE 25 -10 b Grouping 2 A paraphyletic grouping consists of an ancestral species and some, but not all, of the descendants Paraphyletic

LE 25 -10 c Grouping 3 A polyphyletic grouping consists of various species that lack a common ancestor Polyphyletic


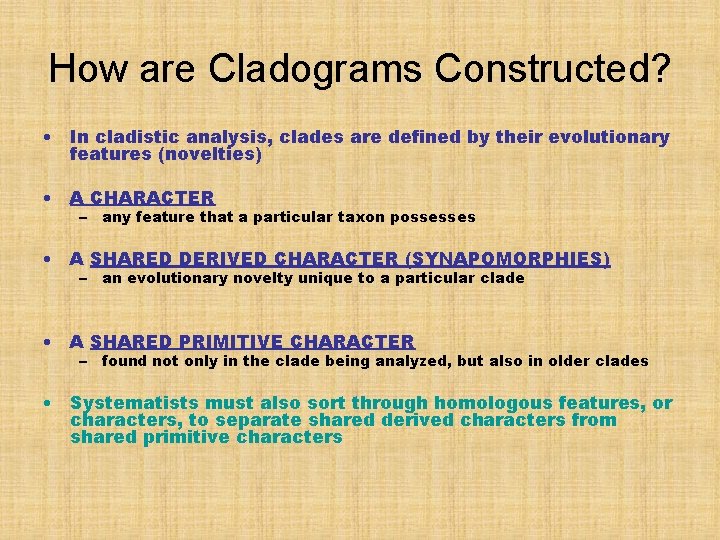
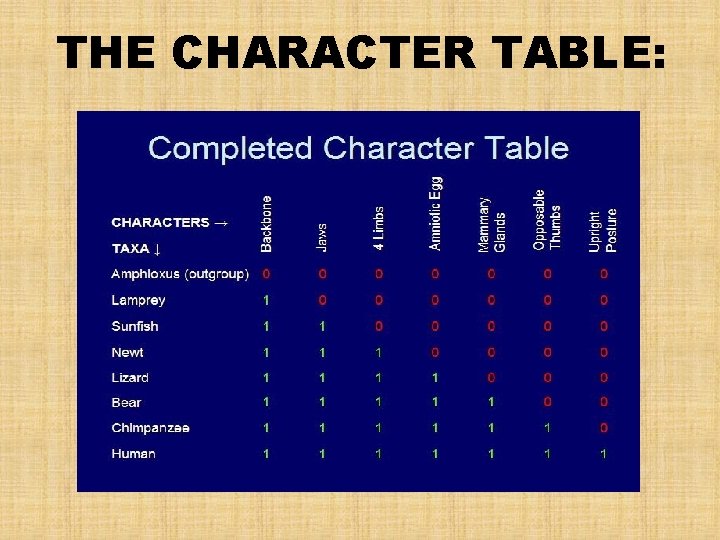
How are Cladograms Constructed? • In cladistic analysis, clades are defined by their evolutionary features (novelties) • A CHARACTER – any feature that a particular taxon possesses • A SHARED DERIVED CHARACTER (SYNAPOMORPHIES) – an evolutionary novelty unique to a particular clade • A SHARED PRIMITIVE CHARACTER – found not only in the clade being analyzed, but also in older clades • Systematists must also sort through homologous features, or characters, to separate shared derived characters from shared primitive characters

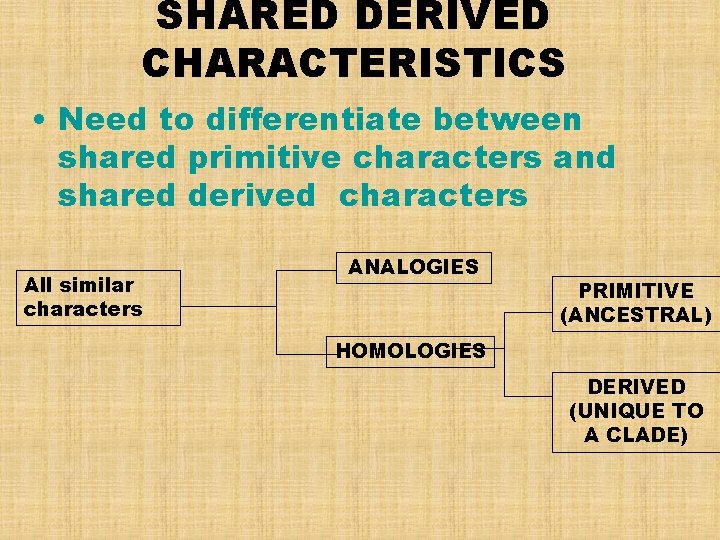
SHARED DERIVED CHARACTERISTICS • Need to differentiate between shared primitive characters and shared derived characters All similar characters ANALOGIES PRIMITIVE (ANCESTRAL) HOMOLOGIES DERIVED (UNIQUE TO A CLADE)

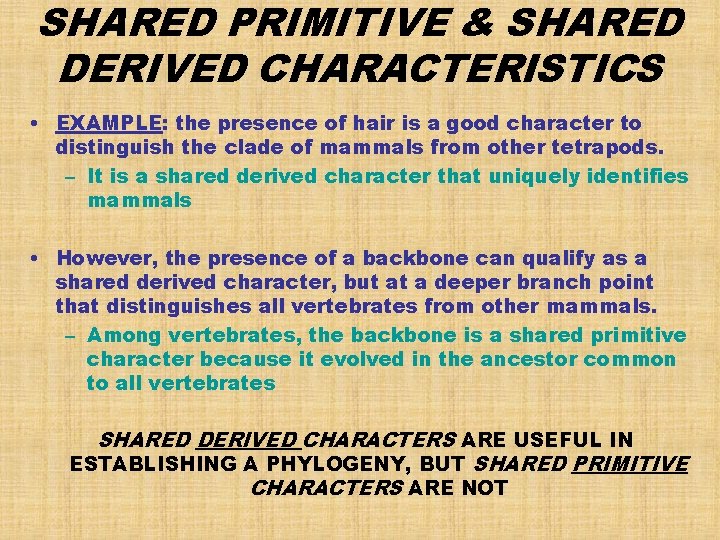
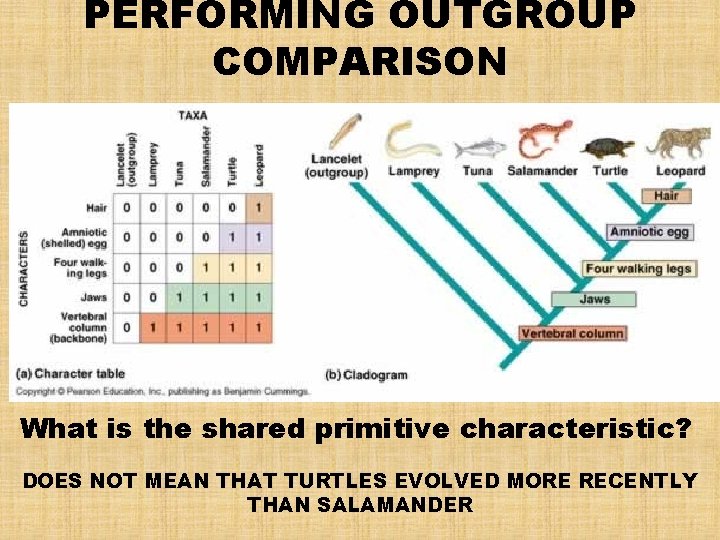
SHARED PRIMITIVE & SHARED DERIVED CHARACTERISTICS • EXAMPLE: the presence of hair is a good character to distinguish the clade of mammals from other tetrapods. – It is a shared derived character that uniquely identifies mammals • However, the presence of a backbone can qualify as a shared derived character, but at a deeper branch point that distinguishes all vertebrates from other mammals. – Among vertebrates, the backbone is a shared primitive character because it evolved in the ancestor common to all vertebrates SHARED DERIVED CHARACTERS ARE USEFUL IN ESTABLISHING A PHYLOGENY, BUT SHARED PRIMITIVE CHARACTERS ARE NOT

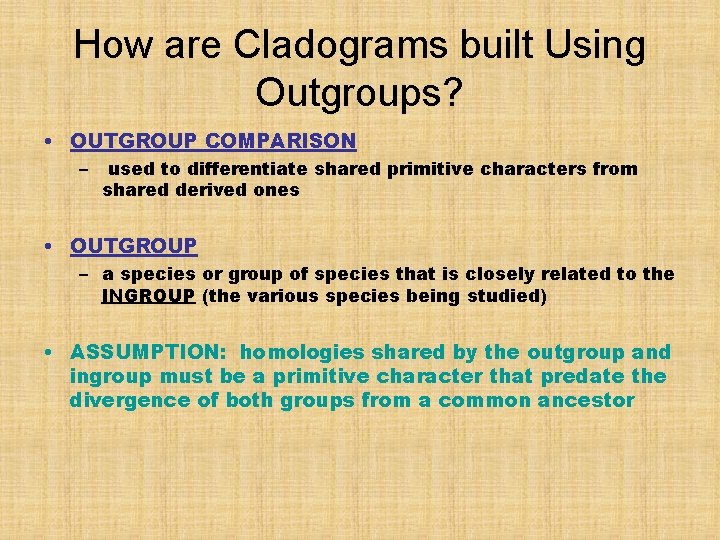
How are Cladograms built Using Outgroups? • OUTGROUP COMPARISON – used to differentiate shared primitive characters from shared derived ones • OUTGROUP – a species or group of species that is closely related to the INGROUP (the various species being studied) • ASSUMPTION: homologies shared by the outgroup and ingroup must be a primitive character that predate the divergence of both groups from a common ancestor

PERFORMING OUTGROUP COMPARISON What is the shared primitive characteristic? DOES NOT MEAN THAT TURTLES EVOLVED MORE RECENTLY THAN SALAMANDER


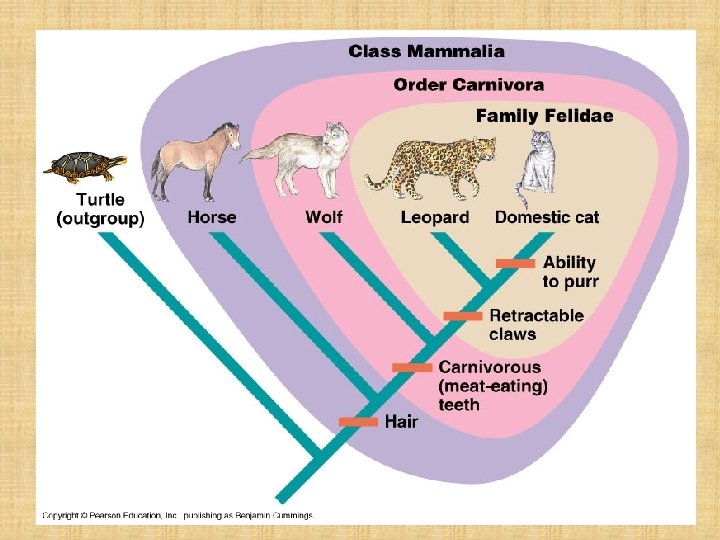
BUILDING A CLADOGRAM

THE CHARACTER TABLE:

THE RESULT:


SIMPLEST EXPLANATION IS NOT THE BEST… Parsimony does not always work, nature does not always take the simplest course

• Much of an organism’s evolutionary history is documented in its genome

What are Gene Duplications and Gene Families? • GENE DUPLICATION – increases the number of genes in the genome, providing more opportunities for evolutionary changes • GENE FAMILIES – groups of related genes within an organism’s genome • Like homologous genes in different species, duplicated genes have a common genetic ancestor – There are two types of homologous genes: ORTHOLOGOUS genes and PARALOGOUS genes

TWO REMARKABLE FACTS ABOUT GENE FAMILIES • All living things share many biochemical and development pathways • The number of genes seems not to have increased at the same rate as phenotypic complexity – Humans have only five times as many genes as yeast, a simple unicellular eukaryote, although we have a large, complex brain and a body that contains more than 200 different types of tissues – Many human genes are more versatile than yeast and can carry out a wide variety of tasks in various body tissues

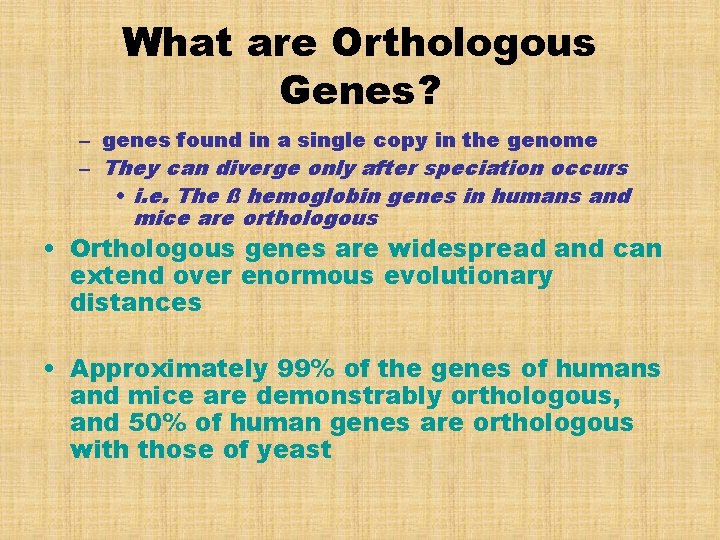
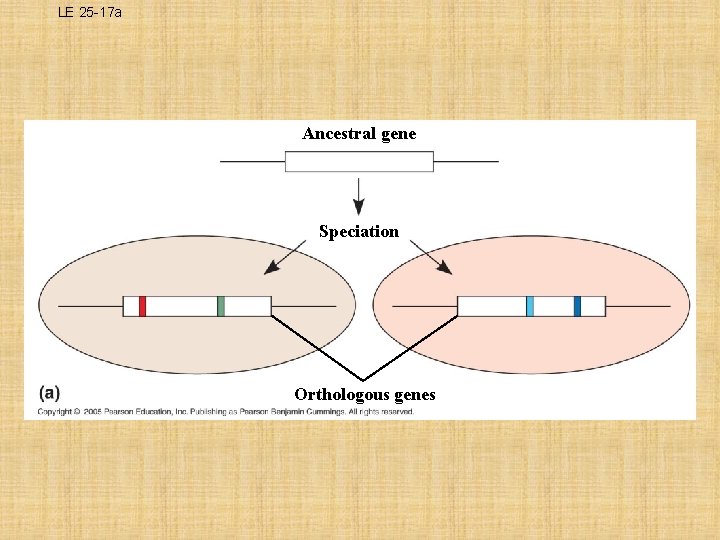
What are Orthologous Genes? – genes found in a single copy in the genome – They can diverge only after speciation occurs • i. e. The ß hemoglobin genes in humans and mice are orthologous • Orthologous genes are widespread and can extend over enormous evolutionary distances • Approximately 99% of the genes of humans and mice are demonstrably orthologous, and 50% of human genes are orthologous with those of yeast

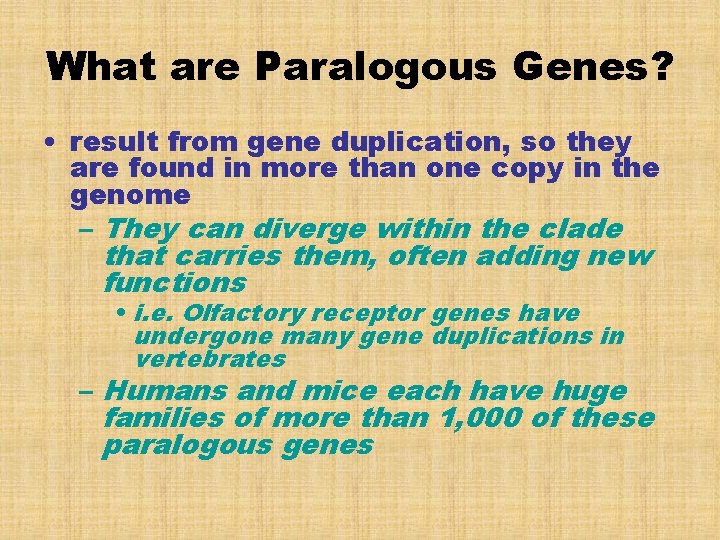
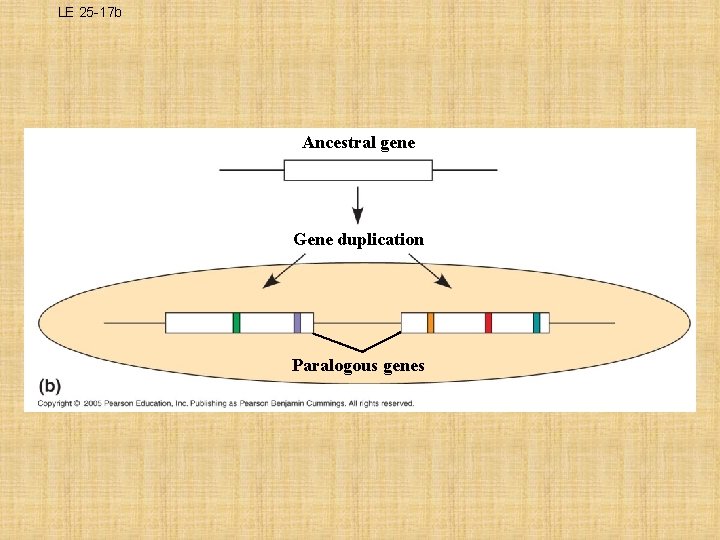
What are Paralogous Genes? • result from gene duplication, so they are found in more than one copy in the genome – They can diverge within the clade that carries them, often adding new functions • i. e. Olfactory receptor genes have undergone many gene duplications in vertebrates – Humans and mice each have huge families of more than 1, 000 of these paralogous genes

LE 25 -17 a Ancestral gene Speciation Orthologous genes

LE 25 -17 b Ancestral gene Gene duplication Paralogous genes

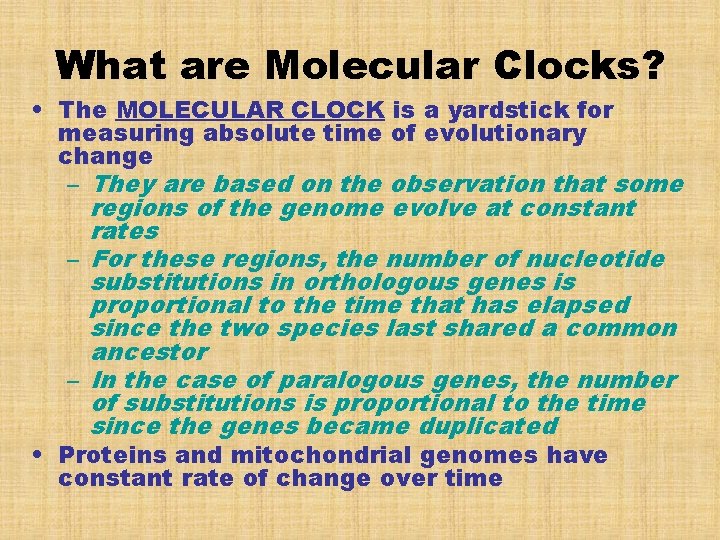
What are Molecular Clocks? • The MOLECULAR CLOCK is a yardstick for measuring absolute time of evolutionary change – They are based on the observation that some regions of the genome evolve at constant rates – For these regions, the number of nucleotide substitutions in orthologous genes is proportional to the time that has elapsed since the two species last shared a common ancestor – In the case of paralogous genes, the number of substitutions is proportional to the time since the genes became duplicated • Proteins and mitochondrial genomes have constant rate of change over time

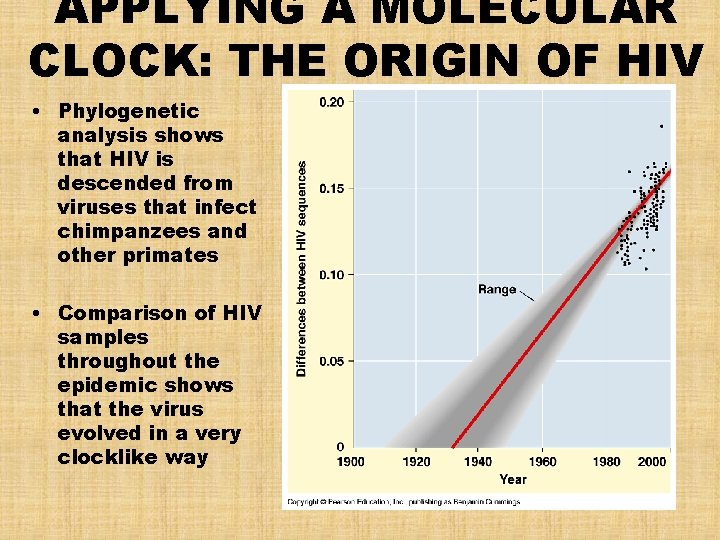
APPLYING A MOLECULAR CLOCK: THE ORIGIN OF HIV • Phylogenetic analysis shows that HIV is descended from viruses that infect chimpanzees and other primates • Comparison of HIV samples throughout the epidemic shows that the virus evolved in a very clocklike way