INTRODUCTION TO DATA FORMATS AND TOOLS DATA FORMATS













![PLINK COMMANDS Command --recode --freq --vcf --keep [file] --remove [file] --extract [file] --exclude [file] PLINK COMMANDS Command --recode --freq --vcf --keep [file] --remove [file] --extract [file] --exclude [file]](https://slidetodoc.com/presentation_image_h/75915b62e0ce84fbc5cd2cd872e84caa/image-20.jpg)






![PLINK FILTERING Command Action --hwe [threshold] Keep variants with HWE p<threshold --missing Compute per-sample PLINK FILTERING Command Action --hwe [threshold] Keep variants with HWE p<threshold --missing Compute per-sample](https://slidetodoc.com/presentation_image_h/75915b62e0ce84fbc5cd2cd872e84caa/image-28.jpg)





- Slides: 33

INTRODUCTION TO DATA FORMATS AND TOOLS DATA FORMATS FOR GWAS AND PLINK SHAUN ARON SYDNEY BRENNER INSTITUTE FOR MOLECULAR BIOSCIENCE UNIVERSITY OF THE WITWATERSRAND

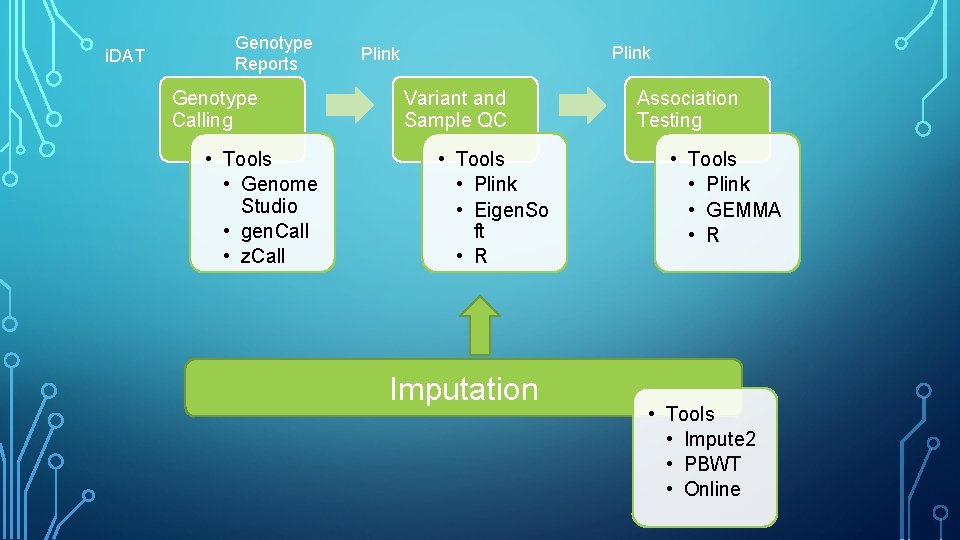
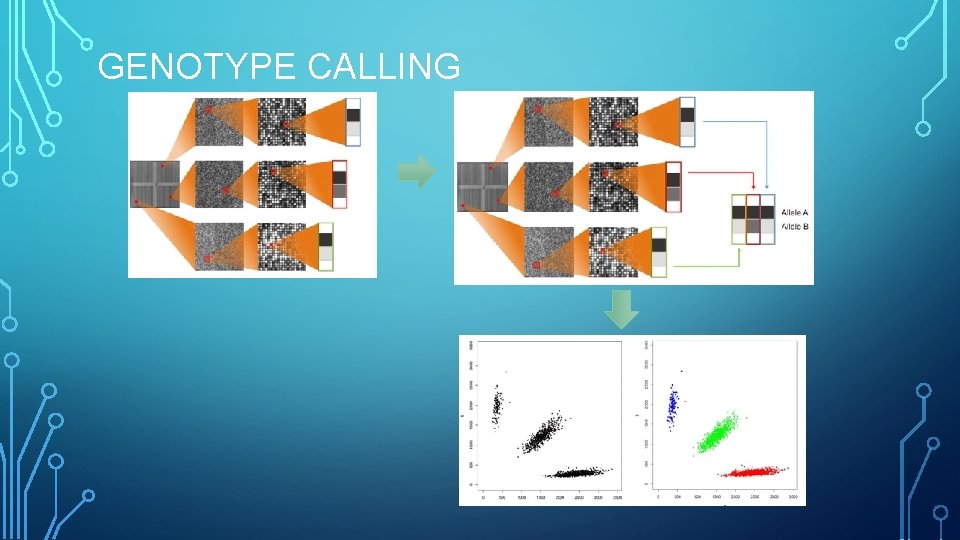
Measure intensities Genotype calling Variant QC Sample QC Association

i. DAT Genotype Reports Genotype Calling • Tools • Genome Studio • gen. Call • z. Call Plink Variant and Sample QC • Tools • Plink • Eigen. So ft • R Imputation Association Testing • Tools • Plink • GEMMA • R • Tools • Impute 2 • PBWT • Online

GENOTYPE CALLING

GENOTYPE CALLS TO PLINK • Some genotyping software exports data in Plink format • There available tools, scripts to convert genotyping reports to plink format or you can do it yourself

PLINK • Plink is standard tool for manipulating and analyzing genotype data • Plink works with standard data formats • • • Has the functionality to convert between different formats Developed and optimized for working with biallelic SNP data Plink online manual • https: //www. cog-genomics. org/plink 2

DATA FORMATS • PED format • • • PED file – Sample/Individual information MAP file – SNV information No header with 6 first defined columns • • • Family ID • Followed by allele calls for the variant in a pairwise fashion – Different encodings Individual ID Paternal ID Maternal ID Sex (1=male, 2=female, other) Phenotype (missing -9, control 1, case 2 or quantitative trait)

PED FORMAT FID IID PATI PHENO Alleles for SNP 1 D MATI SEX D Alleles for SNP 2

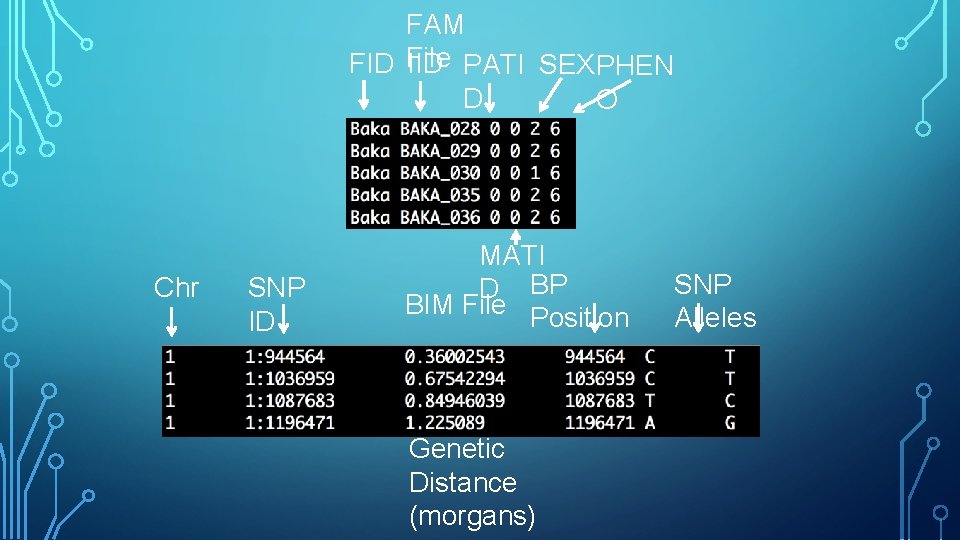
MAP FILE Chr SNP ID BP Position Genetic Distance (morgans)

DATA STORAGE • Plain text format for thousands of samples for millions of SNPs would require a large amount of space for storage • Plink rather works with Binary versions of the PED files • Method to compress and reduce the size of the PED and MAP files

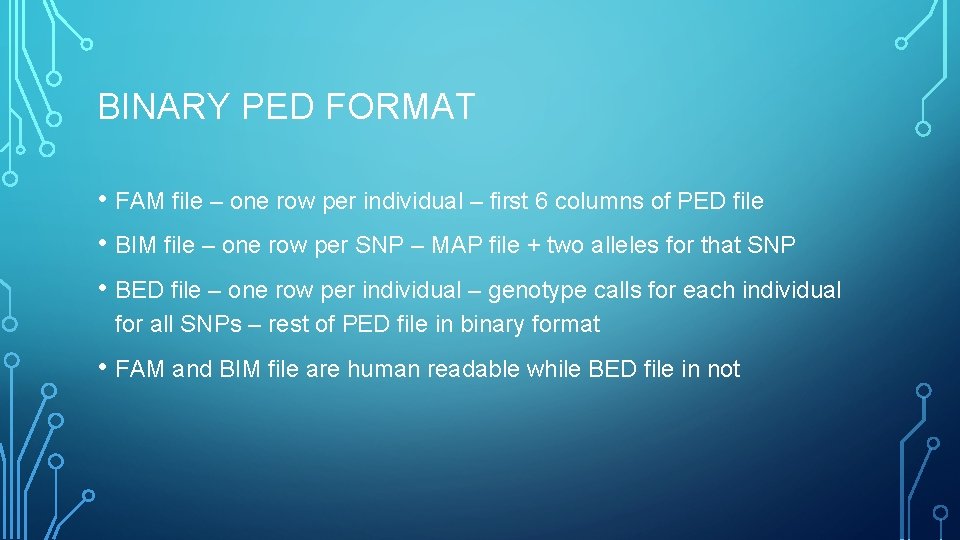
BINARY PED FORMAT • FAM file – one row per individual – first 6 columns of PED file • BIM file – one row per SNP – MAP file + two alleles for that SNP • BED file – one row per individual – genotype calls for each individual for all SNPs – rest of PED file in binary format • FAM and BIM file are human readable while BED file in not

FAM FID File IID PATI SEX PHEN D O Chr SNP ID MATI D BP BIM File Position Genetic Distance (morgans) SNP Alleles

OTHER FORMATS • Plink takes in various other data formats • Able to convert from other formats into Plink format

PLINK BASICS

PLINK BASICS • Command line based • Call Plink using plink command

PLINK BASICS • Flags are used for different operations • Eg. --file used to tell plink the name of the prefix of the input files and • the format Eg. --file hapmap 1 • In your current directory you should have your data in PED format: • hapmap 1. map, hapmap 1. ped • Try it now

PLINK BASICS • Output files have a plink prefix by default. Use --out flag to specify your own name • If you want to explicitly convert to binary format you may use the -make-bed flag

PLINK BASICS • Examine your newly generated files • • Identify what each row and column denotes Remember that you cannot open the. bed file - not human readable • If you are reading in a file in binary PED format use the --bfile flag

RUN THROUGH EXERCISE 2
![PLINK COMMANDS Command recode freq vcf keep file remove file extract file exclude file PLINK COMMANDS Command --recode --freq --vcf --keep [file] --remove [file] --extract [file] --exclude [file]](https://slidetodoc.com/presentation_image_h/75915b62e0ce84fbc5cd2cd872e84caa/image-20.jpg)
PLINK COMMANDS Command --recode --freq --vcf --keep [file] --remove [file] --extract [file] --exclude [file] --pheno [file] Action Transform between formats Generate simple statistics Read in file in VCF format Retain samples in the specified file Remove samples in the specified file Keep SNPs in the specified file Remove SNPS in the specified file Read phenotypes from specified

PLINK FILTERING • May be a need to extract specific parts of a complete dataset • Specific SNPs or Individuals • Can extract either SNPs or Individuals directly on the command line or using a file with a specific format

PLINK FILTERING • For individual filtering you can create a file with the FID and IID of the individuals you want to keep or remove. • For SNP filtering you can create a file with the SNPs IDs you would like to extract or exclude. • In both cases you would most likely generate a new dataset.

PLINK FILTERING Sample File --keep --remove SNP file --extract --exclude

PHENO FILE • Phenotypes can be added to the PED or BIM file • In some instances it is useful to store them in a separate file • Use --pheno flag followed by file with the following format FID IID PHENO

PLINK FILTERING • Another useful flag is the --filter flag • Uses the same file format as the phenotype file • Also has some built is filtering functions • • --filter-cases --filter-controls --filter-males --filter-females

RUN THROUGH EXERCISE 3. 1 – 3. 11

SELECTION BASED ON CRITERIA • Flags defined to select samples/SNPs based on specific criteria • Will come across these again in the QC section of the course
![PLINK FILTERING Command Action hwe threshold Keep variants with HWE pthreshold missing Compute persample PLINK FILTERING Command Action --hwe [threshold] Keep variants with HWE p<threshold --missing Compute per-sample](https://slidetodoc.com/presentation_image_h/75915b62e0ce84fbc5cd2cd872e84caa/image-28.jpg)
PLINK FILTERING Command Action --hwe [threshold] Keep variants with HWE p<threshold --missing Compute per-sample and per-variant missingness --check-sex Check genotype vs phenotype sex based on X chr --genome Compute relatedness based on IBD --maf [threshold] Keep variants with a MAF> threshold --mind [value] Remove individuals with missing data above value --geno [value] Remove SNPs with missing data above value

CRITERIA SELECTION FLAGS • --mind 0. 02 - value of 0. 02 denotes that all individuals with more than 2% • of missing data should be removed --geno 0. 04 – value of 0. 04 indicates that all SNPs with a call rate of less • that 96% should be removed --maf 0. 01 – value of 0. 01 indicates that all SNPs with a minor allele frequency of less than 1% should be removed

GO THROUGH EXERCISE 3. 11 – 3. 22

MERGING DATASETS • Plink has built in tools for merging datasets • Not a straight forward process but useful for population studies • Datasets need to be from the same build, have SNPs called on the same strands etc. • Section 4 deals with how to merge data successfully

ASSOCIATION TESTING • • Plink provides a number of association testing approaches --assoc – assumes there are case/control values in the phenotype column of your PED/BED file or specified phenotype file and runs a simple chi-squared association test --assoc – assumes there are quantitative values in the phenotype column of your PED/BED file or specified phenotype file and runs a regression analysis --linear – runs a linear regression for a quantitative trait allowing for the inclusion of covariates • • Additional options to run adjust for multiple testing and permutation testing This will be covered in more detail in the course

RUN THROUGH SECTIONS 4 AND 5
 Méduce
Méduce Computer data format
Computer data format Mithun subramaniam
Mithun subramaniam Data formats in computer architecture
Data formats in computer architecture Map reduce types
Map reduce types Small group stages formats and culture
Small group stages formats and culture Cutting tools in sewing with pictures and meaning
Cutting tools in sewing with pictures and meaning Montaj maksud
Montaj maksud Rtf to tiff
Rtf to tiff File formats in multimedia
File formats in multimedia Html
Html Input formats in hadoop
Input formats in hadoop Quatrain poems
Quatrain poems Common log format
Common log format Business writing formats
Business writing formats Simplified format of business letter
Simplified format of business letter Radio station music formats
Radio station music formats Difference between letter and memo
Difference between letter and memo Ngs file formats
Ngs file formats Style specification formats
Style specification formats Action recipe
Action recipe Hadoop file formats
Hadoop file formats Style specification formats
Style specification formats Object persistence formats
Object persistence formats Object persistence formats
Object persistence formats Scanf formats
Scanf formats International address formats
International address formats Camtasia 9 codec
Camtasia 9 codec Convert unstructured data to structured
Convert unstructured data to structured Four model approach
Four model approach Typical process description tools include
Typical process description tools include Introduction to data warehousing and data mining
Introduction to data warehousing and data mining Nccer module 4 introduction to power tools
Nccer module 4 introduction to power tools Tool safety quiz
Tool safety quiz