Sequence File Formats Sequence File Formats Different formats


![Fasta • Simplest file format. Easy to parse, easy to use >identifier [optional information] Fasta • Simplest file format. Easy to parse, easy to use >identifier [optional information]](https://slidetodoc.com/presentation_image_h2/ab7445720d1f16c488de797123e24197/image-6.jpg)









- Slides: 18

Sequence File Formats

Sequence File Formats • Different formats for different uses • Competing formats developed in parallel • Some easy to read, some easy to write programs • Don’t have to stick to these formats, but parsers already written! • Most formats are plain text (not. bam files!)

Id’s versus accessions • When people first started, they were using gene names as id’s • But too few gene names, and databases require unique ids • Now have a variety of accession numbers • The simplest id is a number that you increment, as you can (almost) never run out of IDs.

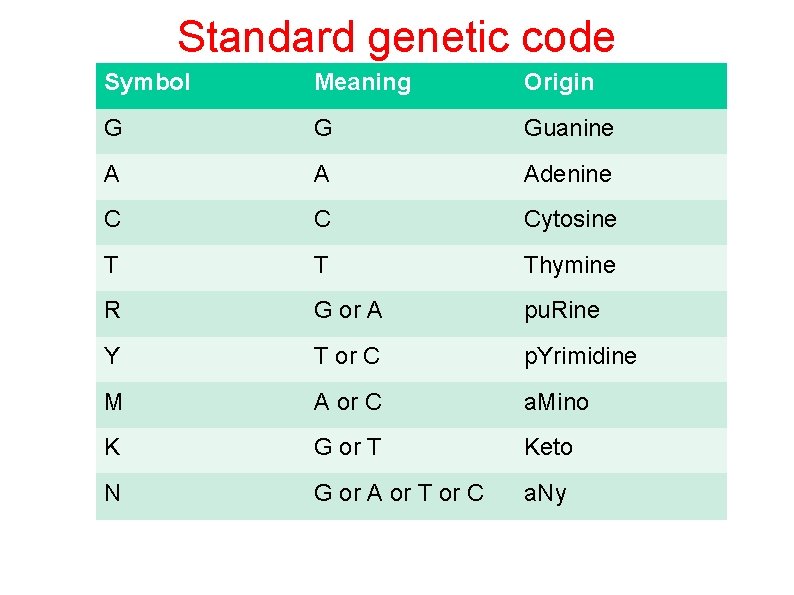
Standard genetic code Symbol Meaning Origin G G Guanine A A Adenine C C Cytosine T T Thymine R G or A pu. Rine Y T or C p. Yrimidine M A or C a. Mino K G or T Keto N G or A or T or C a. Ny

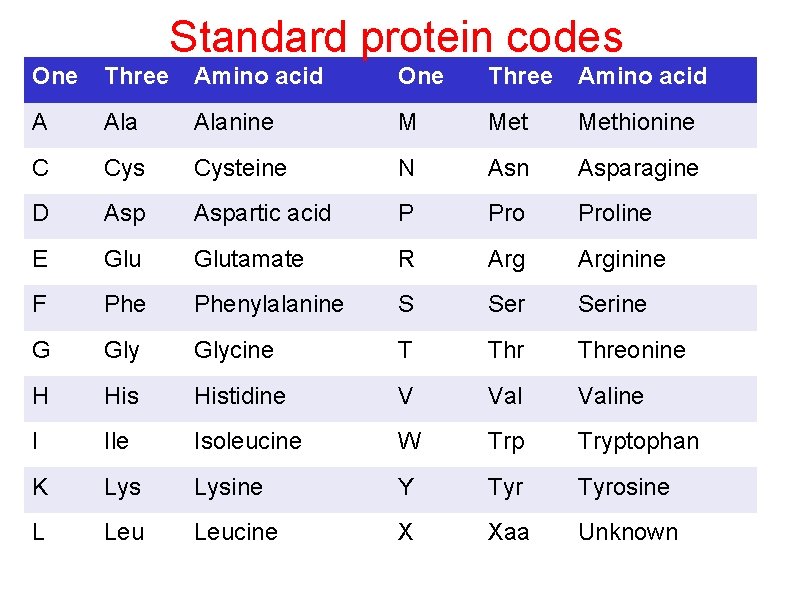
Standard protein codes One Three Amino acid A Alanine M Methionine C Cysteine N Asn Asparagine D Aspartic acid P Proline E Glutamate R Arginine F Phenylalanine S Serine G Glycine T Threonine H Histidine V Valine I Ile Isoleucine W Trp Tryptophan K Lysine Y Tyrosine L Leucine X Xaa Unknown
![Fasta Simplest file format Easy to parse easy to use identifier optional information Fasta • Simplest file format. Easy to parse, easy to use >identifier [optional information]](https://slidetodoc.com/presentation_image_h2/ab7445720d1f16c488de797123e24197/image-6.jpg)
Fasta • Simplest file format. Easy to parse, easy to use >identifier [optional information] ATGACTAGCATCGATCGACTAGCATG ACTGCACTACGACGACAGCAAC >identifier 2 [optional information] ACTAGCTCAGCTAGAGAGCTACGATCAGCACTAC atccgatagcatgacttact. ACGCTAGCATCAGTCATAC AT

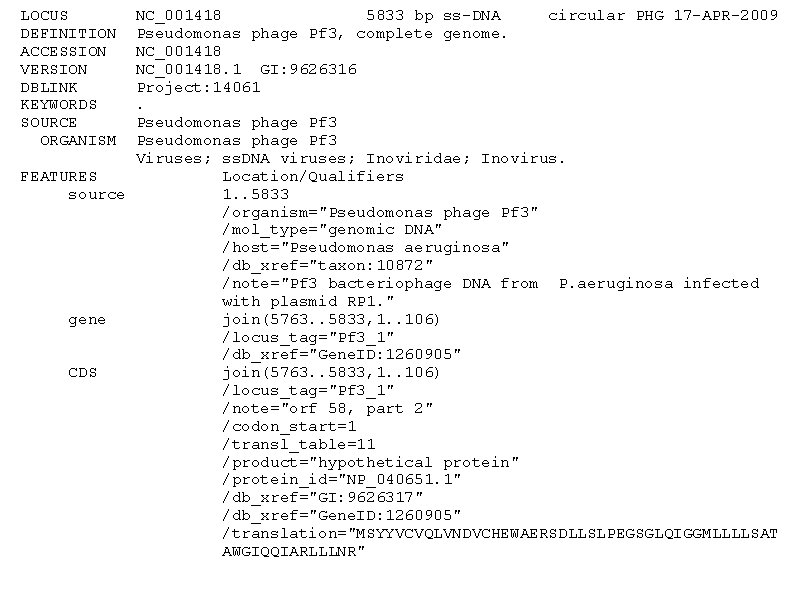
Gen. Bank • More complex, includes detailed information on genes, cds, annotation etc • Human readable • Difficult to parse • Use standard parsers (bioperl, biojava, etc)

LOCUS DEFINITION ACCESSION VERSION DBLINK KEYWORDS SOURCE ORGANISM NC_001418 5833 bp ss-DNA circular PHG 17 -APR-2009 Pseudomonas phage Pf 3, complete genome. NC_001418. 1 GI: 9626316 Project: 14061. Pseudomonas phage Pf 3 Viruses; ss. DNA viruses; Inoviridae; Inovirus. FEATURES Location/Qualifiers source 1. . 5833 /organism="Pseudomonas phage Pf 3" /mol_type="genomic DNA" /host="Pseudomonas aeruginosa" /db_xref="taxon: 10872" /note="Pf 3 bacteriophage DNA from P. aeruginosa infected with plasmid RP 1. " gene join(5763. . 5833, 1. . 106) /locus_tag="Pf 3_1" /db_xref="Gene. ID: 1260905" CDS join(5763. . 5833, 1. . 106) /locus_tag="Pf 3_1" /note="orf 58, part 2" /codon_start=1 /transl_table=11 /product="hypothetical protein" /protein_id="NP_040651. 1" /db_xref="GI: 9626317" /db_xref="Gene. ID: 1260905" /translation="MSYYVCVQLVNDVCHEWAERSDLLSLPEGSGLQIGGMLLLLSAT AWGIQQIARLLLNR"

3241 3301 3361 3421 3481 3541 3601 3661 3721 3781 3841 3901 3961 4021 4081 4141 4201 // LOCUS DEFINITION ACCESSION VERSION DBLINK KEYWORDS SOURCE ORGANISM aggtcctgtt atgagaaaat cggctcttgc atggcgatga atgttggttt gcggaaagca tgagtccctc gcgtatggga ttctccggag taatcagtcc agaggaaaga agcaactcga atggcactct gtgcgtttga tccggaggca caccctaggt tgccctcact ggccttaaga atcctcaatg tcggtcttta cataatattg tcgtaccaac ctactttttg cgatctcata tcctagggta gaatgtcgtg tttcgcagaa ccgagttcgt atgtgacagt cgcttacgca catcatgtgg cgaaggctct tagcaatact cctccca tcacccaagg ggtaatggct tgcgaattac ccatcagacg aagaagaaaa ggcgttgacg ctggttttga tatcctgtat cctgatggat aatcgcggtt gacgaatatg gagttcccct caccgagaac ataccgtgca atcctaaaaa taaactaacc gcatcttgcc ataccttcga tgggcttacg cgtgcagtcc cgttttctag tcacaccttt accagatgta acaccaagta acggtgatgg gggttcggcg gttcgtatct ttaacgggtc ggttacctac gtagtcgtgt tggggtagcg ttctcaaaag agatggtacc gcttgagtcg accgtcagat tctagttgaa tggaccgttc ctacatacgt tcgttgggcc tagacgttac tgccctcgtc tgtgccgatg ctacgagcta gctggtcgttatcagt cctggctccc cctgggaggg agagagtgaa gtcattactt cttatatttg gttacggtct gttttctcct cgagagtcgt cgccgtatag acaattgacg cttccggaaa ggatctgtct attatagaca tggtcgttgc ggttccactg gatgccgtaa tacggggatt gtgcattatg ggctctgctt NC_003301 3192 bp ds-RNA linear PHG 23 -AUG-2008 Pseudomonas phage phi 8 segment S, complete sequence. NC_003301. 1 GI: 17736965 Project: 14731. Pseudomonas phage phi 8 Viruses; ds. RNA viruses; Cystoviridae; Cystovirus.

GFF 3 • Tab separated format • Easy to parse • Attributes are tag/value pairs separated by “; ” • Columns: 1. Contig 2. Source database 3. Feature type 4. Start 5. Stop 6. Score 7. Strand 8. Phase 9. Attributes

ASN. 1 • Developed as computer readable form of Gen. Bank • Not widely used

ASN. 1 seq { id { local id 1 }, descr { title "" }, inst { repr raw, mol aa, length 131, topology linear, { seq-data iupacaa "TSPASIRPPAGPSSRPAMVSSRRTRPSPPGPRRPTGRPCCSAAPRRPQA TGGWKTCSGTCTTSTSTRHRGRSGWSARTTTAACLRASRKSMRAACSRSAGSRPNRFA PTLMSSCITSTTGPPAWAGDRSHE" } } , seq { id { local id 1 }, descr { title "" }, inst { repr raw, mol aa, length 131, topology linear, { seq-data iupacaa "TSPASIRPPAGPSSR-----RPSPPGPRRPTGRPCCSAAPRRPQAT GGWKTCSGTCTTSTSTRHRGRSGW-----RASRKSMRAACSRSAGSRPNRFAPTL MSSCITSTTGPPAWAGDRSHE" } }

Base calling Need to be sure which base you have identified Depends on the technology Each machine includes software Phred is an historical package developed by at U. Washington Phred scores are probability that the base is correct

Quality values Phred 10: 1 x 101 chance that the base is wrong Phred 20: 1 x 102 chance that the base is wrong Phred 30: 1 x 103 chance that the base is wrong Phred 40: 1 x 104 chance that the base is wrong Phred 99: the base is correct! Fastq scores are the score + 33 then converted to ascii text

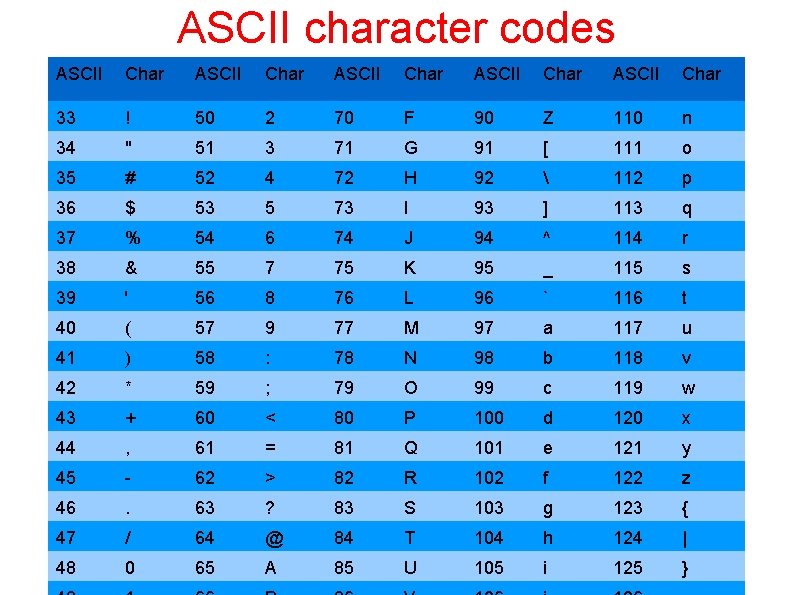
Fast. Q • Based on fasta format • Contains information about the quality of the sequence • Quality comes from sequencing machines! • Four lines per sequence: • Line starting @ = identifier line before the sequence • DNA sequence • Line starting + = identifier line before the quality scores • String = quality scores as ASCII + 33

ASCII character codes ASCII Char ASCII Char 33 ! 50 2 70 F 90 Z 110 n 34 " 51 3 71 G 91 [ 111 o 35 # 52 4 72 H 92 112 p 36 $ 53 5 73 I 93 ] 113 q 37 % 54 6 74 J 94 ^ 114 r 38 & 55 7 75 K 95 _ 115 s 39 ' 56 8 76 L 96 ` 116 t 40 ( 57 9 77 M 97 a 117 u 41 ) 58 : 78 N 98 b 118 v 42 * 59 ; 79 O 99 c 119 w 43 + 60 < 80 P 100 d 120 x 44 , 61 = 81 Q 101 e 121 y 45 - 62 > 82 R 102 f 122 z 46 . 63 ? 83 S 103 g 123 { 47 / 64 @ 84 T 104 h 124 | 48 0 65 A 85 U 105 i 125 }

fastq @SRR 014849. 1 EIXKN 4201 CFU 84 length=93 DNA sequence GGGGGGGGCTTTTTTTGGAACCGA AAGGGTTTTGAATTTCAAACCCTTTTCGGTTTCCAA CCTTCCAAAGCAATGCCAATA +SRR 014849. 1 EIXKN 4201 CFU 84 length=93 3+&$#""""""7 F@71, '"; C? , B; ? 6 B; : E A 1 EA 5’ 9 B: ? : #9 EA 0 D@2 EA 5': >5? : %A; A 8 A; ? 9 B; D@/=<? 7=9<2 A 8== Quality scores Note: Illumina has a format of fastq that is not compatible with everyone else’s format!

How to convert fastq to fasta prinseq-lite. pl -fastq input. fastq -out_format 2 https: //edwards. sdsu. edu/research/fastq-tofasta/