NGS File formats Raw data from various vendors







- Slides: 17

NGS File formats • Raw data from various vendors => various formats • Different quality metrics (some more stringent than others) • As data analysis proceeds, end up with even more formats: – – Gen. Bank formats (SRA) Alignments are in SAM/BAM Genome Browser formats (wig, bed, gff, etc) Variants in vcf files (SNPs, indels, etc)

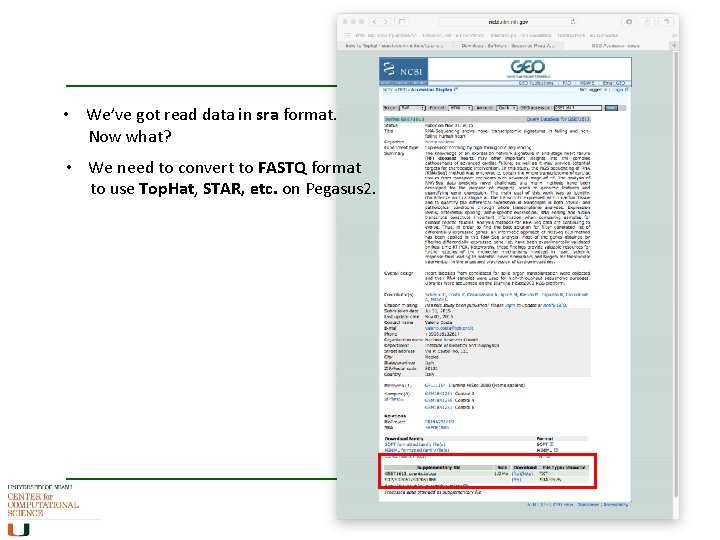
What, if there is no Galaxy, …? • Looking for Sequence reads data (SRA) • http: //www. ncbi. nlm. nih. gov/sra , http: //www. ebi. ac. uk/ena for example:

• We’ve got read data in sra format. Now what? • We need to convert to FASTQ format to use Top. Hat, STAR, etc. on Pegasus 2.

SRA toolkit http: //www. ncbi. nlm. nih. gov/Traces/sra/? view=software

SRA toolkit fastq-dump –X 5 –Z SRR 390728 fastq-dump –I --split-files SRR 390728 fastq-dump --split-files –fasta 60 SRR 390728

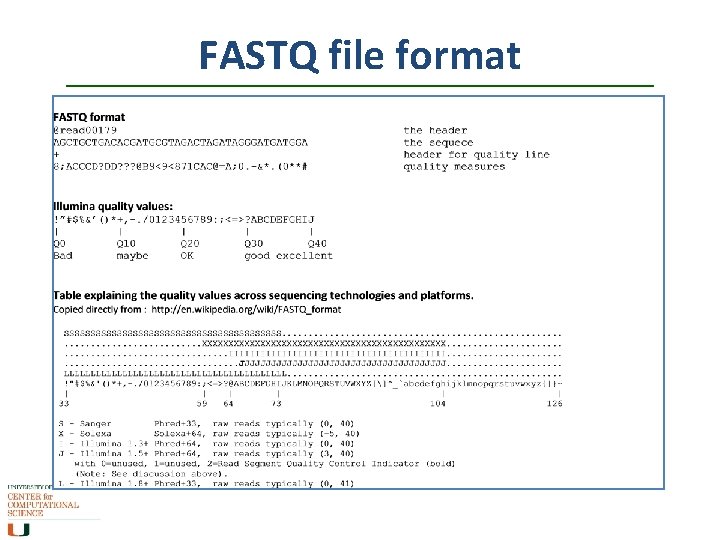
FASTQ file format

Working on pegasus 2 • We’ve got our fastq file(s). • To align the reads with the reference genome we will use Top. Hat on pegasus 2 • transfer data through • scp yourfile. fastq <user>@pegasus 2 -gw. ccs. miami. edu: ~/. into your home directory • scp yourfiel. fastq <user>@pegasus 2 gw. ccs. miami. edu: /scratch/<user> into scratch directory

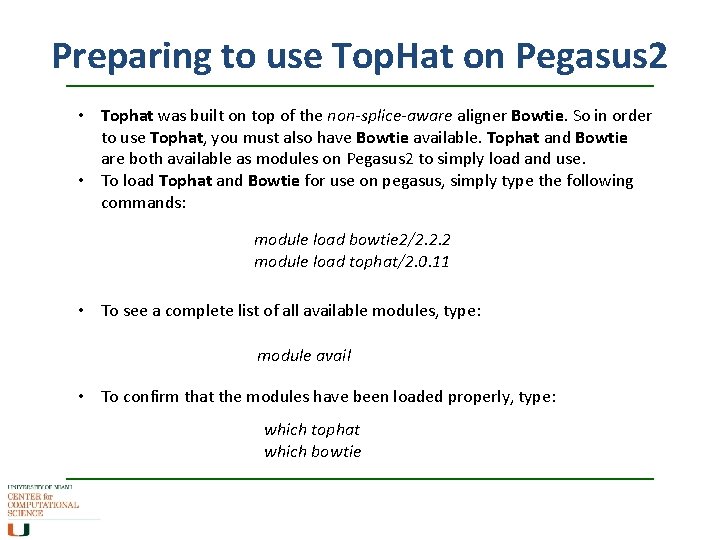
Preparing to use Top. Hat on Pegasus 2 • Tophat was built on top of the non-splice-aware aligner Bowtie. So in order to use Tophat, you must also have Bowtie available. Tophat and Bowtie are both available as modules on Pegasus 2 to simply load and use. • To load Tophat and Bowtie for use on pegasus, simply type the following commands: module load bowtie 2/2. 2. 2 module load tophat/2. 0. 11 • To see a complete list of all available modules, type: module avail • To confirm that the modules have been loaded properly, type: which tophat which bowtie

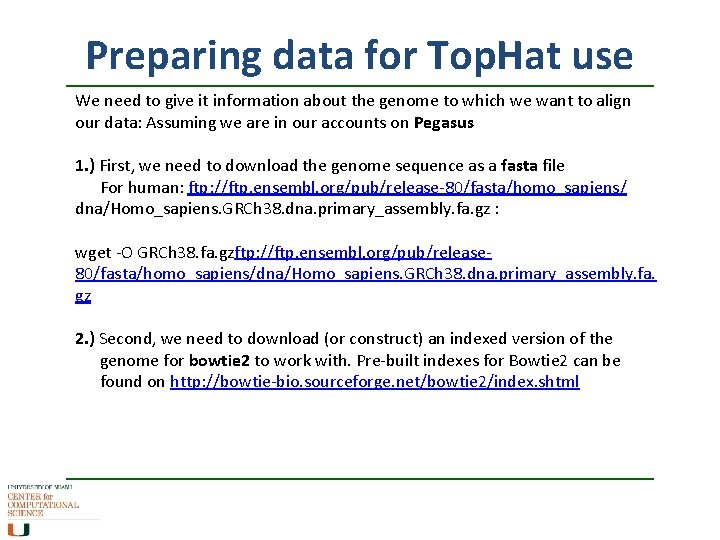
Preparing data for Top. Hat use We need to give it information about the genome to which we want to align our data: Assuming we are in our accounts on Pegasus 1. ) First, we need to download the genome sequence as a fasta file For human: ftp: //ftp. ensembl. org/pub/release-80/fasta/homo_sapiens/ dna/Homo_sapiens. GRCh 38. dna. primary_assembly. fa. gz : wget -O GRCh 38. fa. gzftp: //ftp. ensembl. org/pub/release 80/fasta/homo_sapiens/dna/Homo_sapiens. GRCh 38. dna. primary_assembly. fa. gz 2. ) Second, we need to download (or construct) an indexed version of the genome for bowtie 2 to work with. Pre-built indexes for Bowtie 2 can be found on http: //bowtie-bio. sourceforge. net/bowtie 2/index. shtml

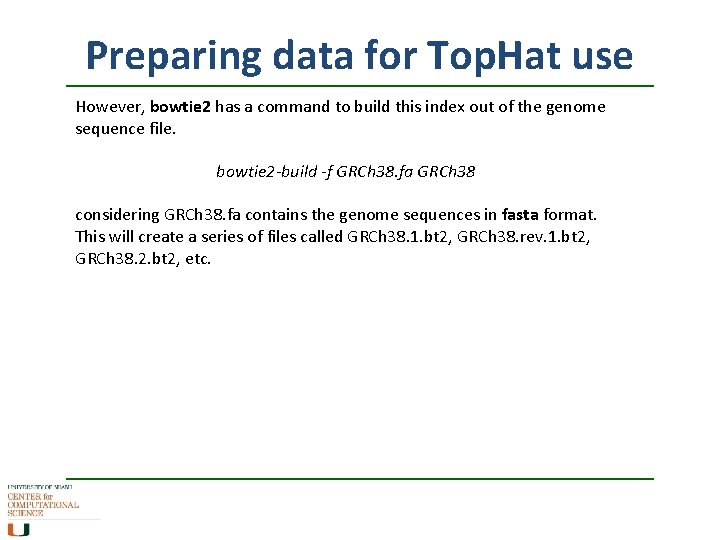
Preparing data for Top. Hat use However, bowtie 2 has a command to build this index out of the genome sequence file. bowtie 2 -build -f GRCh 38. fa GRCh 38 considering GRCh 38. fa contains the genome sequences in fasta format. This will create a series of files called GRCh 38. 1. bt 2, GRCh 38. rev. 1. bt 2, GRCh 38. 2. bt 2, etc.

Preparing data for Top. Hat use 3. ) Third, we need to download an annotation file containing all the known genes and transcripts for this genome. This third step is technically optional, but it helps to improve the accuracy of splice junction calling and is generally recommended. We are also going to need this file when we quantify the transcripts present, so might as well use it now too. We need to give it information about the genome to which we want to align our data: Genome annotation: Human: ftp: //ftp. ensembl. org/pub/release 80/gtf/homo_sapiens/Homo_sapiens. GRCh 38. 80. gtf. gz) wget -O GRCh 38. gtfftp: //ftp. ensembl. org/pub/release 80/gtf/homo_sapiens/Homo_sapiens. GRCh 38. 80. gtf. gz) We are ready to run Top. Hat!

Running Top. Hat on Pegasus 2 To run a job on Pegasus 2 in the background we need to create a shell script (http: //ccs. miami. edu/hpc/support/faq/): which shell? job name #!/bin/bash file for stderr #BSUB –J Tophat_job 1 file for stdout #BSUB –e Tophat_job 1. err #BSUB –o Tophat_job 1. out number of cores #BSUB –n 4 queue, more info http: //ccs. miami. edu/hpc/doc/pegasus 2 -queues/ #BSUB -q general #BSUB -W 72: 00 time allocation # Your actual commands for the job are going to be placed here.

Running Top. Hat The complete list of Top. Hat parameters and their official descriptions are http: //ccb. jhu. edu/software/tophat/manual. shtml Optional parameters: -G … specify a genomic annotation to use -o … locate the output directory Required parameters: - 'base name' of the genome sequence/index data so that it can go and find it. - fastq files to use

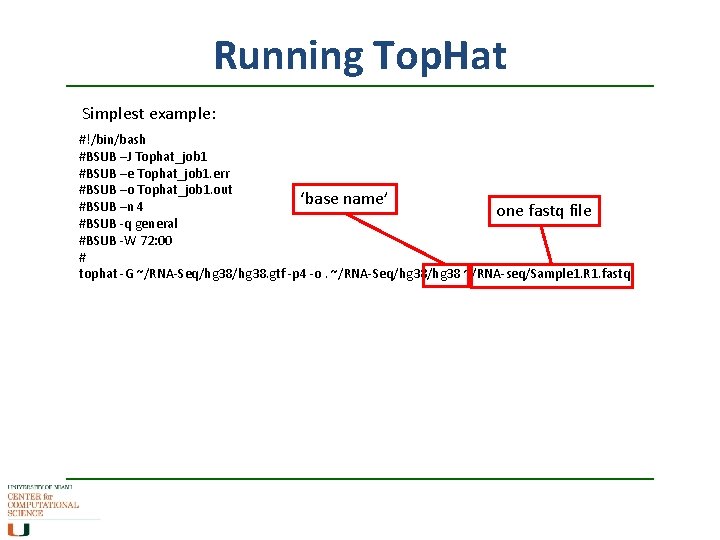
Running Top. Hat Simplest example: #!/bin/bash #BSUB –J Tophat_job 1 #BSUB –e Tophat_job 1. err #BSUB –o Tophat_job 1. out ‘base name’ #BSUB –n 4 one fastq file #BSUB -q general #BSUB -W 72: 00 # tophat -G ~/RNA-Seq/hg 38. gtf -p 4 -o. ~/RNA-Seq/hg 38 ~/RNA-seq/Sample 1. R 1. fastq

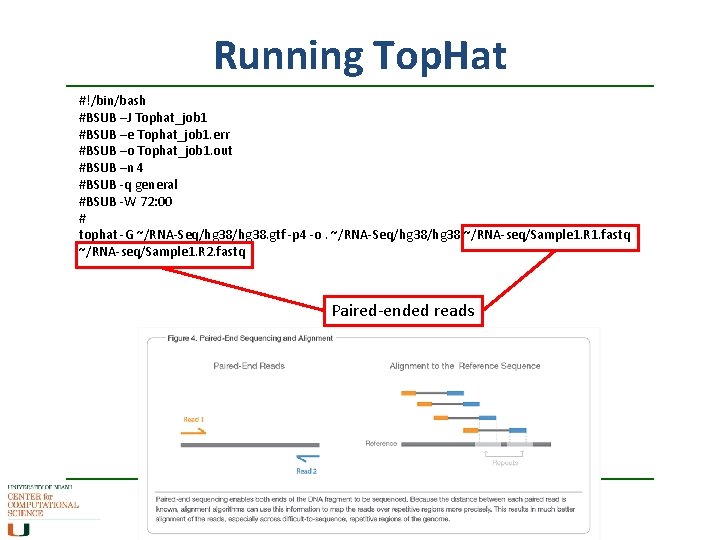
Running Top. Hat #!/bin/bash #BSUB –J Tophat_job 1 #BSUB –e Tophat_job 1. err #BSUB –o Tophat_job 1. out #BSUB –n 4 #BSUB -q general #BSUB -W 72: 00 # tophat -G ~/RNA-Seq/hg 38. gtf -p 4 -o. ~/RNA-Seq/hg 38 ~/RNA-seq/Sample 1. R 1. fastq ~/RNA-seq/Sample 1. R 2. fastq Paired-ended reads

Running Top. Hat #!/bin/bash #BSUB –J Tophat_job 1 #BSUB –e Tophat_job 1. err #BSUB –o Tophat_job 1. out #BSUB –n 4 #BSUB -q general #BSUB -W 72: 00 # tophat -G ~/RNA-Seq/hg 38. gtf -p 4 -o. ~/RNA-Seq/hg 38 ~/Fast. QFiles/Sample 1. Lane 1. R 1. fastq, ~/Fast. QFiles/Sample 1. Lane 2. R 1. fastq, ~/Fast. QFiles/Sample 1. multiple fastq files Lane 3. R 1. fastq tophat -G ~/RNA-Seq/hg 38. gtf -p 4 -o. ~/RNA-Seq/hg 38 ~/Fast. QFiles/Sample 1. Lane 1. R 1. fastq, ~/Fast. QFiles/Sample 1. Lane 2. R 1. fastq, ~/Fast. QFiles/Sample 1. Lane 3. R 1. fastq ~/Fast. QFiles/Sample 1. Lane 1. R 2. fastq, ~/Fast. QFiles/Sample 1. Lane 2. R 2. fastq, ~/Fast. QFiles/Sample 1. Lane 3. R 2. fastq multiple fastq files wit paired-ended reads

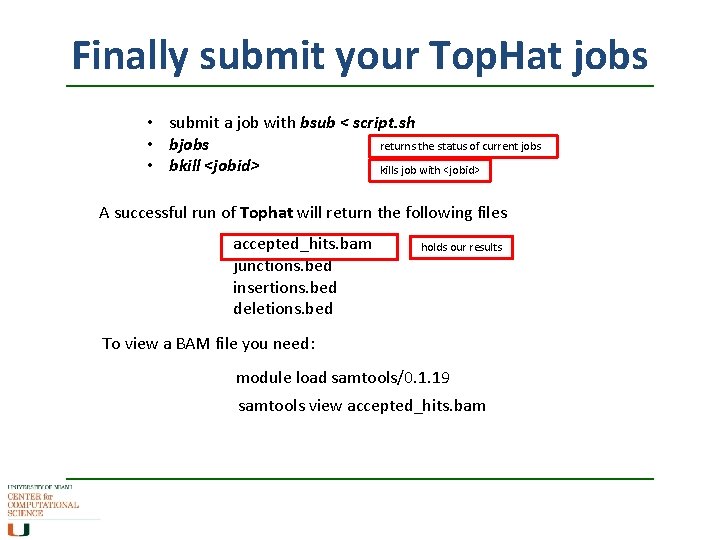
Finally submit your Top. Hat jobs • submit a job with bsub < script. sh • bjobs returns the status of current jobs • bkill <jobid> kills job with <jobid> A successful run of Tophat will return the following files accepted_hits. bam junctions. bed insertions. bed deletions. bed holds our results To view a BAM file you need: module load samtools/0. 1. 19 samtools view accepted_hits. bam