In Search of the Elusive Enhancer Jim Kent


- Slides: 18

In Search of the Elusive Enhancer Jim Kent University of California Santa Cruz

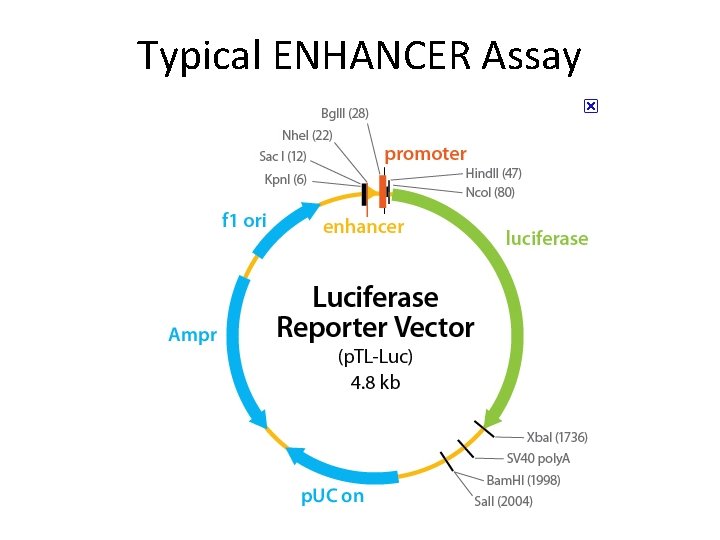
Typical ENHANCER Assay

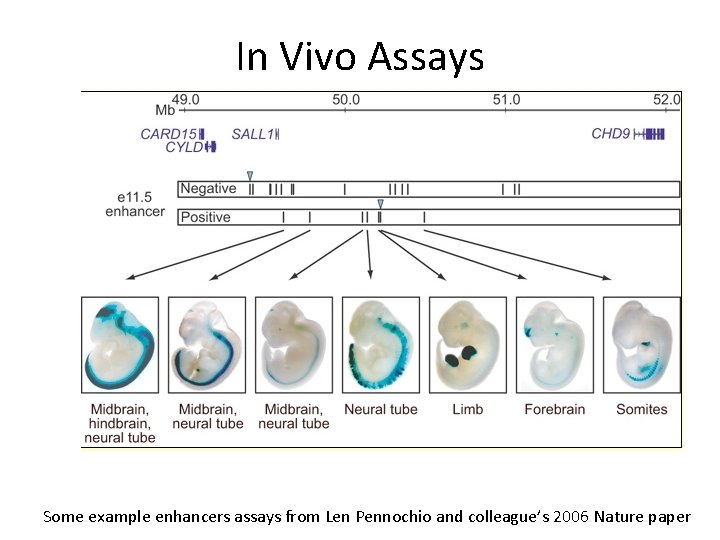
In Vivo Assays Some example enhancers assays from Len Pennochio and colleague’s 2006 Nature paper

Genomics ENHANCER Screens • Conservation – weak but something • P 300 - relatively specific. Sensitivity? • H 3 K 4 Me 1 – association with P 300 and DNAse found by Bing Ren and others, but very broad. • DNAse Hypersensitivity – good but hits promoters more strongly than enhancers • Segmentation algorithms on *various* histone modifications - like H 3 K 4 Me 1 complicated by enhancer itself being near nucleosome free.

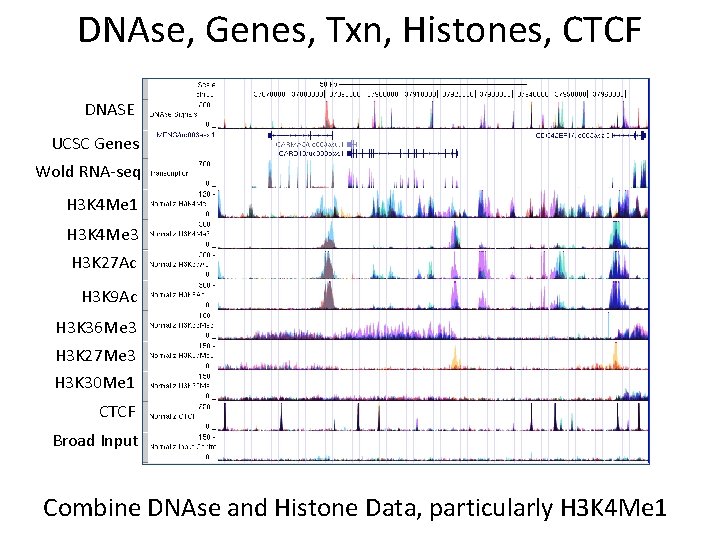
DNAse, Genes, Txn, Histones, CTCF DNASE UCSC Genes Wold RNA-seq H 3 K 4 Me 1 H 3 K 4 Me 3 H 3 K 27 Ac H 3 K 9 Ac H 3 K 36 Me 3 H 3 K 27 Me 3 H 3 K 30 Me 1 CTCF Broad Input Combine DNAse and Histone Data, particularly H 3 K 4 Me 1

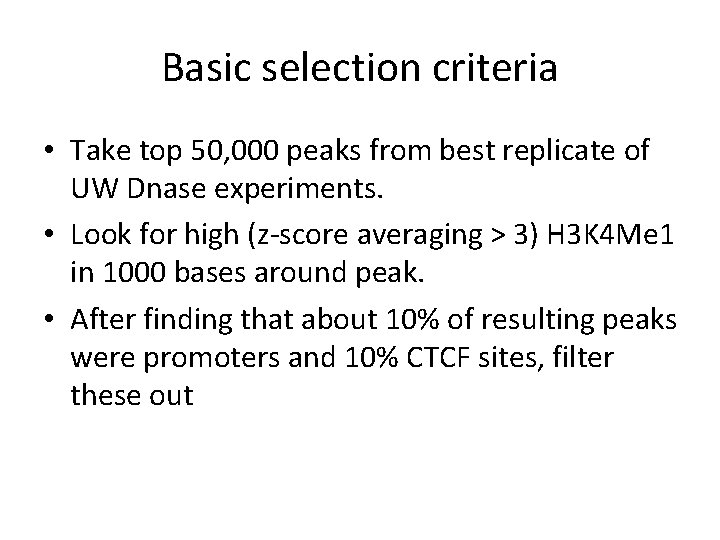
Basic selection criteria • Take top 50, 000 peaks from best replicate of UW Dnase experiments. • Look for high (z-score averaging > 3) H 3 K 4 Me 1 in 1000 bases around peak. • After finding that about 10% of resulting peaks were promoters and 10% CTCF sites, filter these out

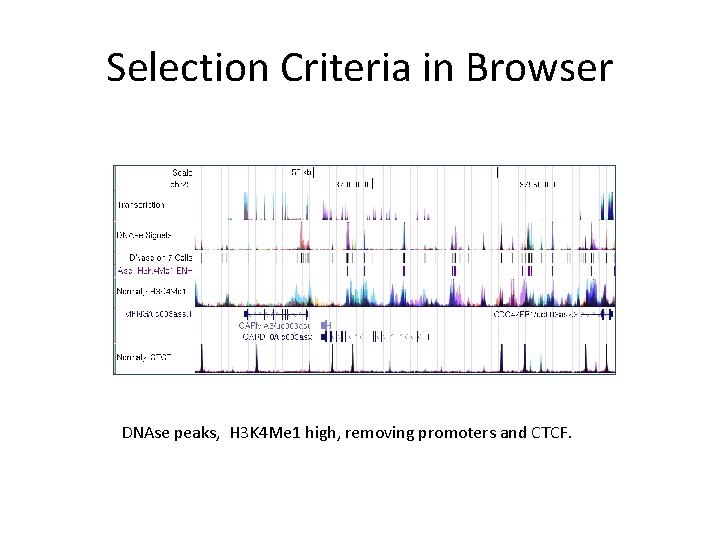
Selection Criteria in Browser DNAse peaks, H 3 K 4 Me 1 high, removing promoters and CTCF.

Enhancer-like properties • Though not included in selection criteria, H 3 K 27 Ac levels very high in our regions. • Very high overlap with P 300 sites – enrichment of 656 x and coverage of 49% of P 300 sites when remove promoters and CTCF from P 300. • “Activity” correlated with nearby promoters in a way that is inhibited by CTCF.

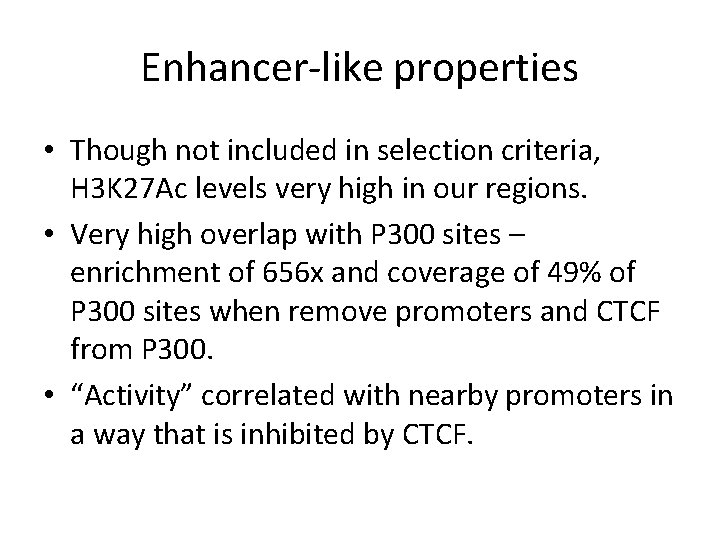
Average signals around features DNAse + H 3 K 4 Me 1 – Pro - CTCF Start of second coding exon Active promoters in GM 12878 CTCF Sites Blue H 3 K 4 Me 1, purple H 3 K 27 Ac, red H 3 K 4 Me 3, orange DNAse, brown RNA-seq, black conserv.

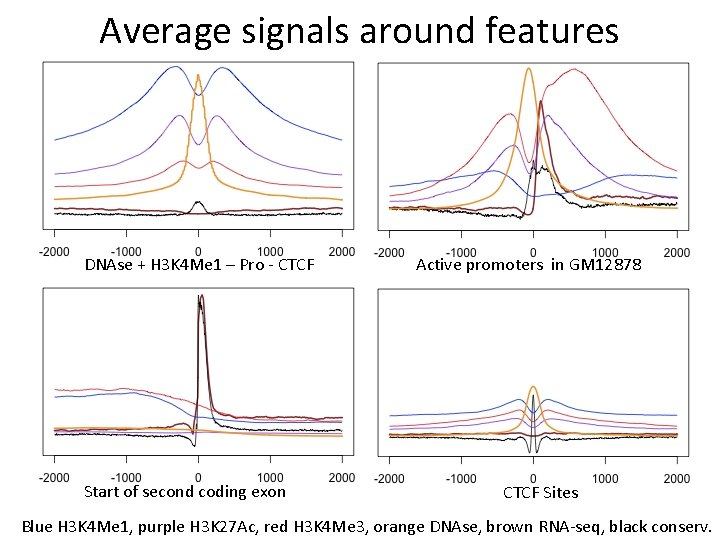
Comparing vs. Chrom. HMM DNAse + H 3 K 4 Me 1 – Pro - CTCF Jason Ernst’s Chromatin HMM: 60, 000 Stringent Enhancers. Blue H 3 K 4 Me 1, purple H 3 K 27 Ac, red H 3 K 4 Me 3, orange DNAse, brown RNA-seq, black conserv.

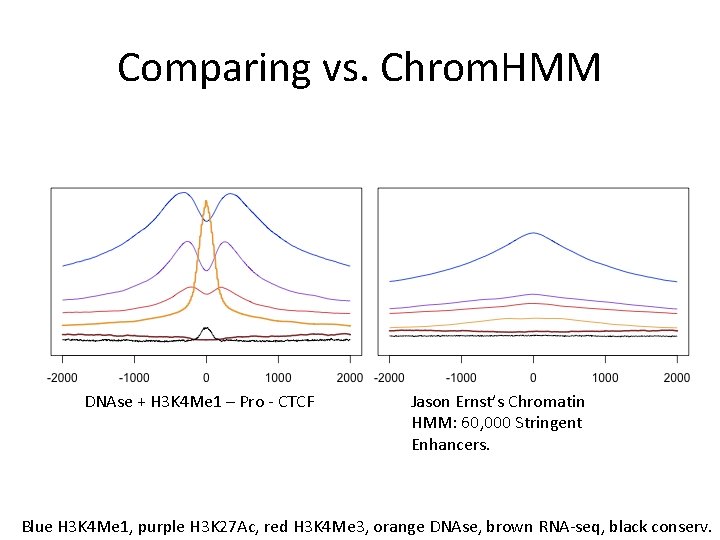
Comparing to P 300 3867 IDR filtered P 300 sites on Gm 12878 from HAIB Top 3867 scoring H 3 K 4 Me 1+DNAse enhancers Blue H 3 K 4 Me 1, purple H 3 K 27 Ac, red H 3 K 4 Me 3, orange DNAse, brown RNA-seq, black conserv.

In search of Enhancer “activity” • Wanted to do correlations of enhancer activity with nearby promoter activity. • Can use RNA signal from gene for promoter activity. • Tried DNAse, H 3 K 4 Me 1 levels as an enhancer activity, but surprisingly H 3 K 27 Ac worked better.

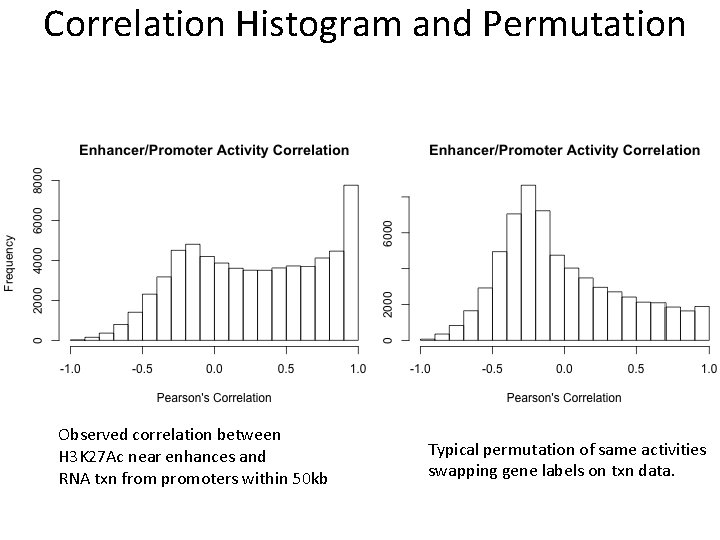
Correlation Histogram and Permutation Observed correlation between H 3 K 27 Ac near enhances and RNA txn from promoters within 50 kb Typical permutation of same activities swapping gene labels on txn data.

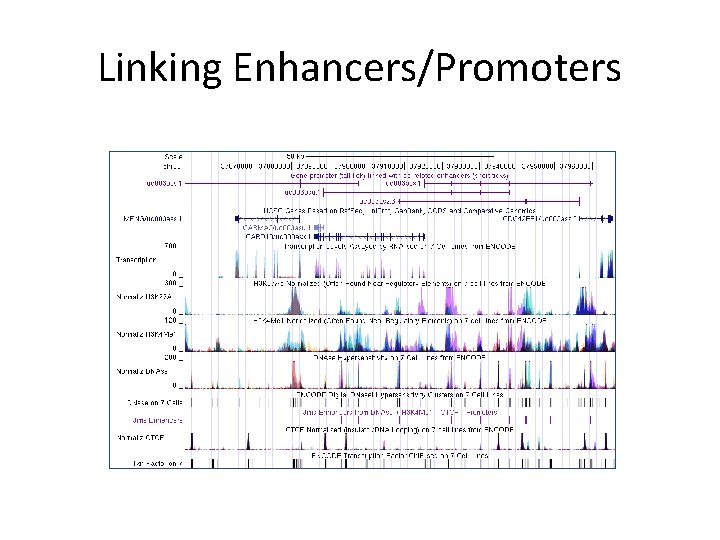
Linking Enhancers/Promoters

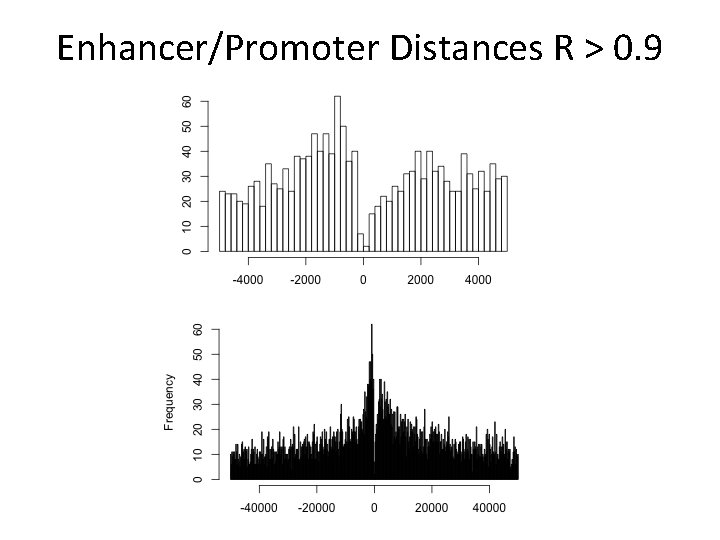
Enhancer/Promoter Distances R > 0. 9

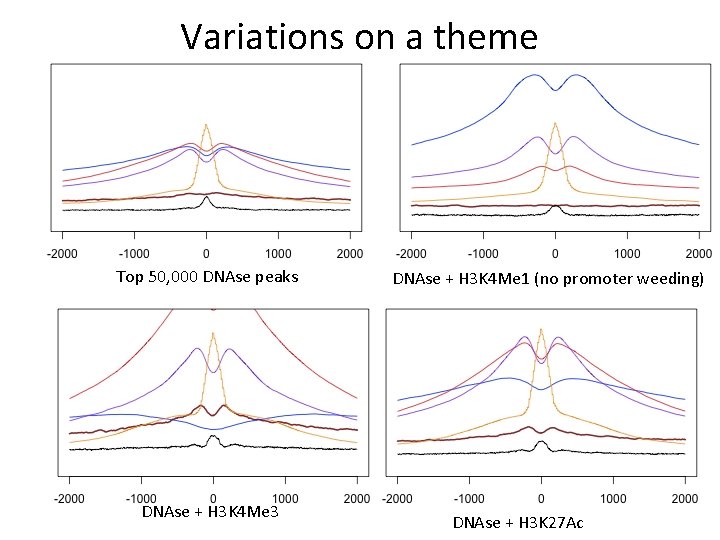
Variations on a theme Top 50, 000 DNAse peaks DNAse + H 3 K 4 Me 3 DNAse + H 3 K 4 Me 1 (no promoter weeding) DNAse + H 3 K 27 Ac

Future Work • Doing luciferase assays on random sample of predicted enhancers • Using a HMM that looks for histone-dnasehistone to sharpen peak calling. • Consider H 3 K 27 Ac+ H 3 K 4 Me 3 - DNAse+ as another worthwhile chromatin based scheme • Comparing transcription factors found at predicted enhancers vs. promoters, on enhancers in one cell line vs. another. • Analyzing non-enhancer, non-promoter, non. CTCF DNAse peaks.

Acknowledgements • • Bernstein lab – Histone and CTCF data. Stamatoyannopoulos lab – DNAse data. Wold lab – RNA-seq data. Myers lab – P 300 data on GM 12878 Anshul – peak calling and IDR on P 300 Bing Ren – for previous work on H 3 K 4 Me 1 NHGRI – funding ENCODE consortium, AWG, DCC for all their hard work.
 Public opinion is both an elusive and fragile commodity.
Public opinion is both an elusive and fragile commodity. Seiser meaning
Seiser meaning Enhancer promoter
Enhancer promoter Enhancer trap drosophila
Enhancer trap drosophila Enhancer vs promoter
Enhancer vs promoter Spectral pitch display adobe audition
Spectral pitch display adobe audition Contoh flavor enhancer
Contoh flavor enhancer Enhancer
Enhancer Contoh flavor sintetis
Contoh flavor sintetis Enhancer promoter
Enhancer promoter Dan dial molecular enhancer
Dan dial molecular enhancer Flavour enhancer 627 and 631 side effects
Flavour enhancer 627 and 631 side effects Advantage and disadvantage of binary search tree
Advantage and disadvantage of binary search tree Federated search vs distributed search
Federated search vs distributed search Local search vs global search
Local search vs global search Which search strategy is called as blind search
Which search strategy is called as blind search Blind search dan heuristic search
Blind search dan heuristic search Linear search vs binary search
Linear search vs binary search Informed and uninformed search in artificial intelligence
Informed and uninformed search in artificial intelligence