GENE INTERACTIONS GENE INTERACTIONS Consider two independent genes
















- Slides: 68

GENE INTERACTIONS

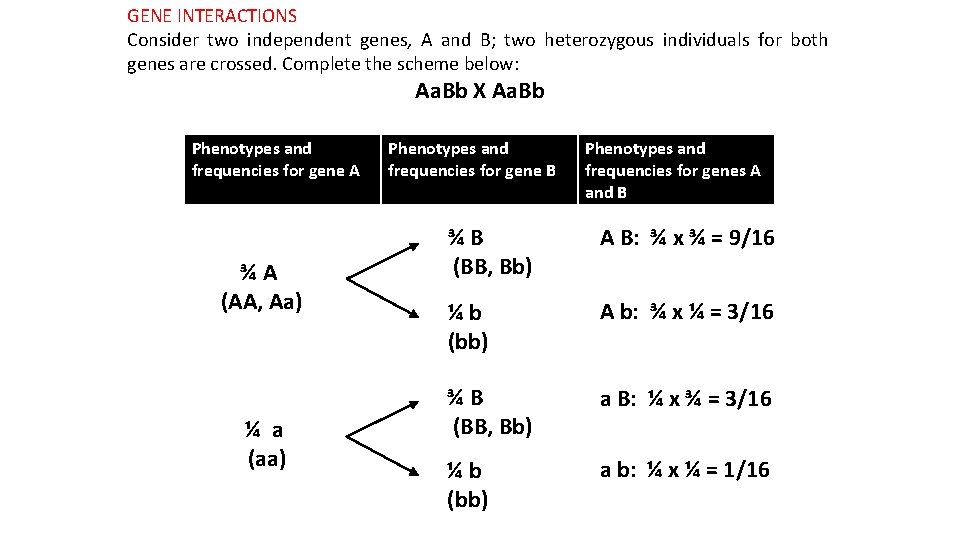
GENE INTERACTIONS Consider two independent genes, A and B; two heterozygous individuals for both genes are crossed. Complete the scheme below: Aa. Bb X Aa. Bb Phenotypes and for gene A frequencies ¾A (AA, Aa) ¼ a (aa) Phenotypes and frequencies for gene B Phenotypes and frequencies for genes A and B ¾B (BB, Bb) A B: ¾ x ¾ = 9/16 ¼b (bb) A b: ¾ x ¼ = 3/16 ¾B (BB, Bb) a B: ¼ x ¾ = 3/16 ¼b (bb) a b: ¼ x ¼ = 1/16

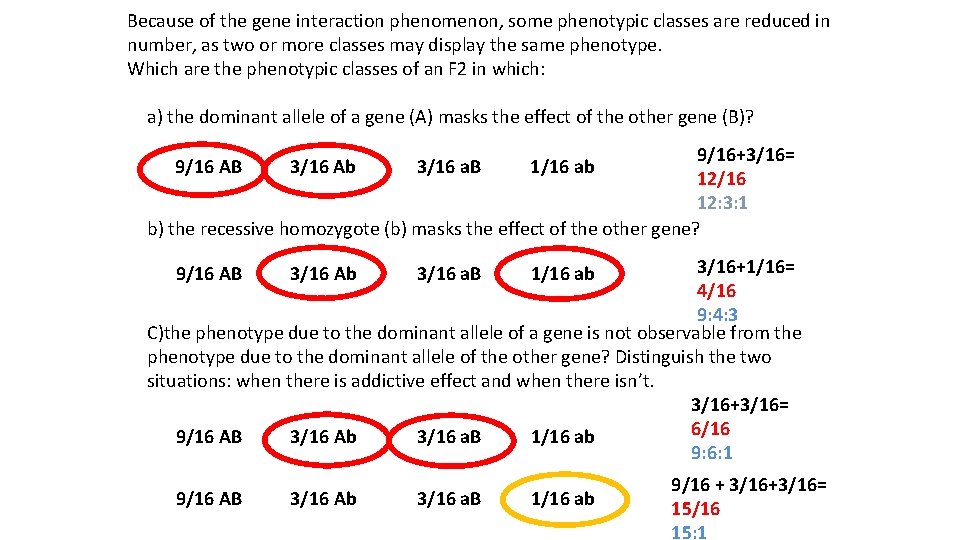
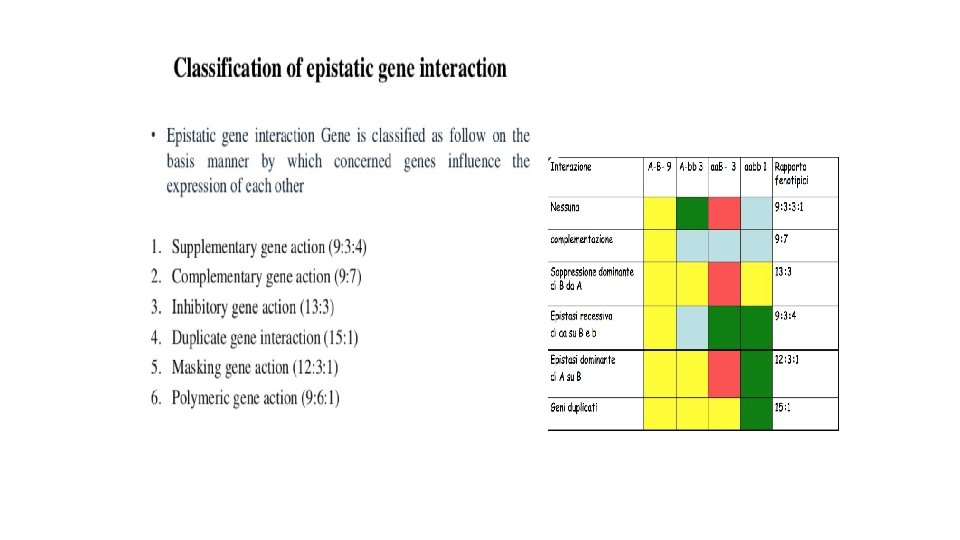
Because of the gene interaction phenomenon, some phenotypic classes are reduced in number, as two or more classes may display the same phenotype. Which are the phenotypic classes of an F 2 in which: a) the dominant allele of a gene (A) masks the effect of the other gene (B)? 9/16+3/16= 12/16 12: 3: 1 b) the recessive homozygote (b) masks the effect of the other gene? 9/16 AB 3/16 Ab 3/16 a. B 1/16 ab 3/16+1/16= 4/16 9: 4: 3 C)the phenotype due to the dominant allele of a gene is not observable from the phenotype due to the dominant allele of the other gene? Distinguish the two situations: when there is addictive effect and when there isn’t. 3/16+3/16= 6/16 9/16 AB 3/16 Ab 3/16 a. B 1/16 ab 9: 6: 1 9/16 AB 3/16 Ab 3/16 a. B 1/16 ab 9/16 + 3/16+3/16= 15/16 15: 1

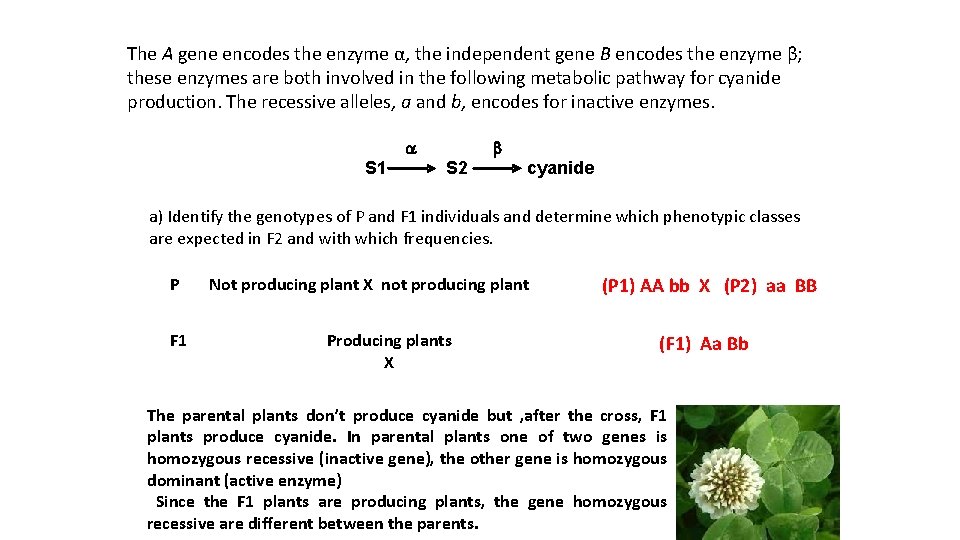
The A gene encodes the enzyme α, the independent gene B encodes the enzyme β; these enzymes are both involved in the following metabolic pathway for cyanide production. The recessive alleles, a and b, encodes for inactive enzymes. a S 1 b S 2 cyanide a) Identify the genotypes of P and F 1 individuals and determine which phenotypic classes are expected in F 2 and with which frequencies. P F 1 Not producing plant X not producing plant Producing plants X (P 1) AA bb X (P 2) aa BB (F 1) Aa Bb The parental plants don’t produce cyanide but , after the cross, F 1 plants produce cyanide. In parental plants one of two genes is homozygous recessive (inactive gene), the other gene is homozygous dominant (active enzyme) Since the F 1 plants are producing plants, the gene homozygous recessive are different between the parents.

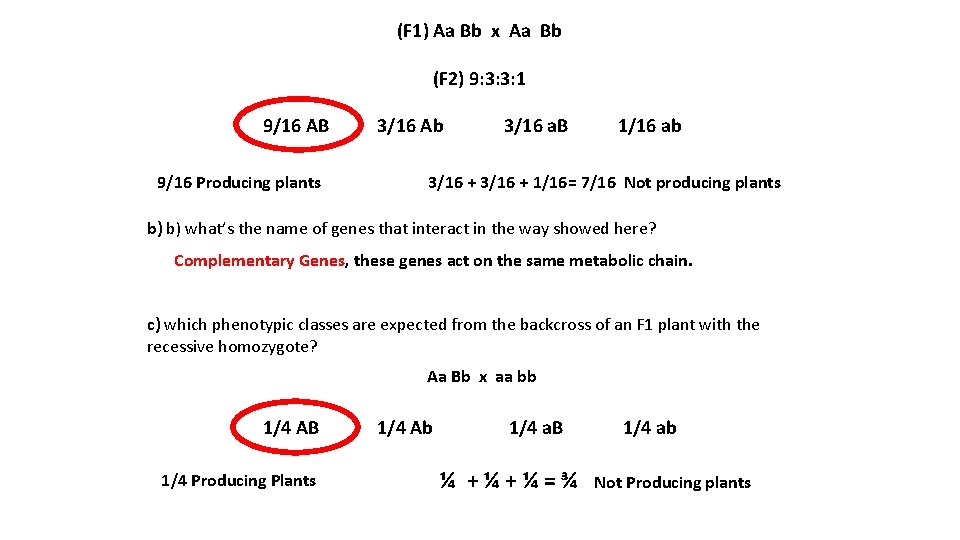
(F 1) Aa Bb x Aa Bb (F 2) 9: 3: 3: 1 9/16 AB 9/16 Producing plants 3/16 Ab 3/16 a. B 1/16 ab 3/16 + 1/16= 7/16 Not producing plants b) b) what’s the name of genes that interact in the way showed here? Complementary Genes, these genes act on the same metabolic chain. c) which phenotypic classes are expected from the backcross of an F 1 plant with the recessive homozygote? Aa Bb x aa bb 1/4 AB 1/4 Producing Plants 1/4 Ab 1/4 a. B ¼ +¼+¼=¾ 1/4 ab Not Producing plants

The two independent genes P and C control two different consecutive steps in the same metabolic pathway. S 1 P S 2 C Yellow fruit The dominant alleles P and C encode for active enzymes, while the recessive alleles, p and c, encode for inactive enzymes. Which phenotype is expected for individuals with the following genotype (intermediate substrates are colourless): a) PPcc This plant produces the enzyme encoded by gene P, but not produces the other one (gene C). Metabolic chain is blocked at step S 2. Phenotype: white melon b) Pp. Cc The plant has both enzymes thus the metabolic chain works until the end. Phenotype: yellow melon. c) pp. CC The plant hasn’t the enzyme encoded by gene P. It produces the enzyme encoded by the gene C but it doesn’t convert S 1 in S 2. Phenotype white melon. d) Ppcc The plant produces the first enzyme but not the second one. Then the metabolic chain is blocked at S 1 step. Phenotype white melon.

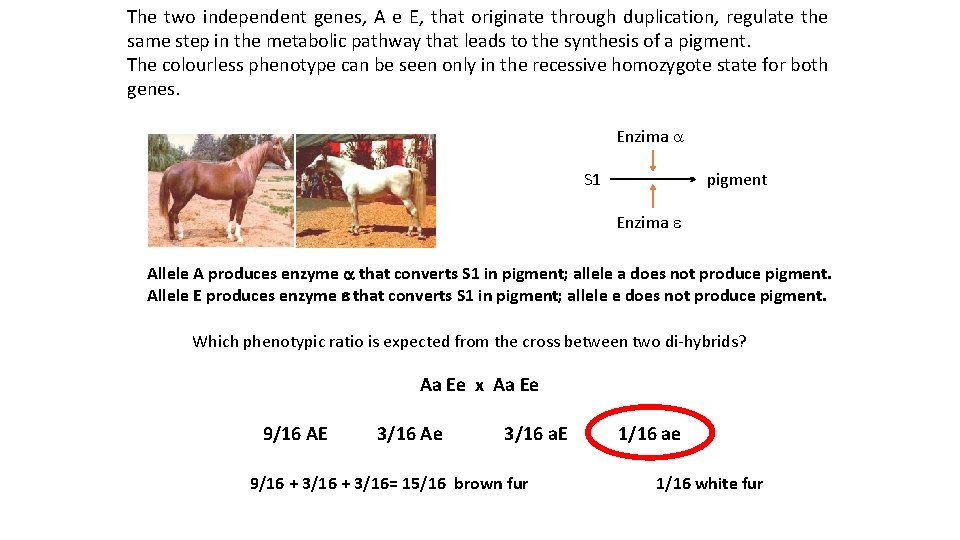
The two independent genes, A e E, that originate through duplication, regulate the same step in the metabolic pathway that leads to the synthesis of a pigment. The colourless phenotype can be seen only in the recessive homozygote state for both genes. Enzima a S 1 pigment Enzima e Allele A produces enzyme a that converts S 1 in pigment; allele a does not produce pigment. Allele E produces enzyme e that converts S 1 in pigment; allele e does not produce pigment. Which phenotypic ratio is expected from the cross between two di-hybrids? Aa Ee x Aa Ee 9/16 AE 3/16 Ae 3/16 a. E 9/16 + 3/16= 15/16 brown fur 1/16 ae 1/16 white fur

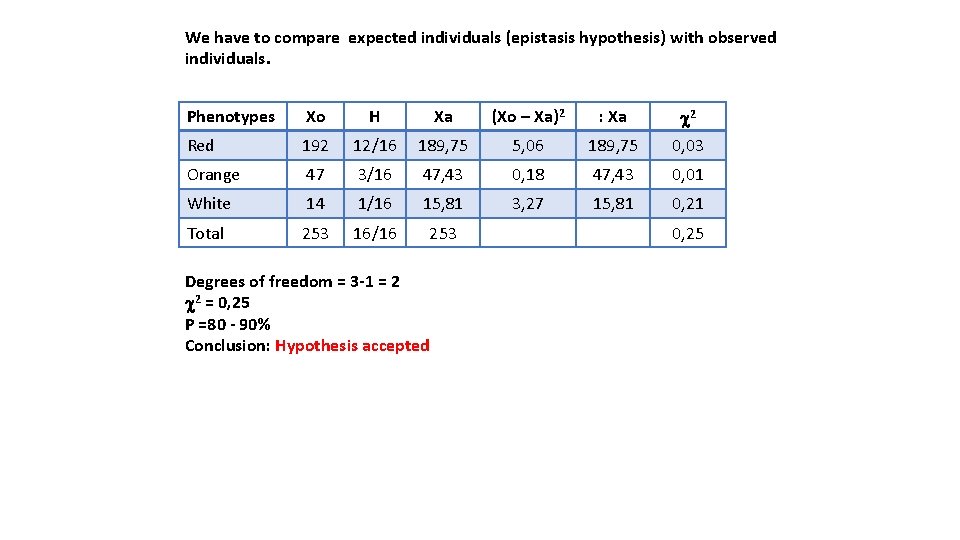
Two pure lines of pepper are crossed: first line with Red fruits and the second one with orange fruits. The plants of F 1 produce only red peppers, but crossing two individuals of F 1 we obtain: 192 RED FRUITS, 47 ORANGE FRUITS, 14 WHITE FRUITS Pure lines = they pass the same alleles to all offsprings and so they are homozygous We know that the colour fruit is controlled by 2 genes (A and B), which kind of interaction is present? Determine the genotypes and hypothesize a possible action on metabolic chain. (P 1) RED FRUITS X (P 2) ORANGE FRUITS AA bb aa BB (F 1) RED FRUITS Aa Bb The dominant allele A is epistatic on gene B F 2: 192 red fruits 9/16 AB + 3/16 Ab 47 orange fruits 3/16 a. B 14 white fruits 1/16 ab When the plant has an allele of A the phenotype related to gene B is not shown. Phenotypes AB and Ab produce red fruits When A is homozygous recessive, the plant show the B phenotype: if it has dominant allele B the fruits will be orange, if it is homozygous bb the fruits will be white. 192+47+14= 253 (: 16= 15. 8)

We have to compare expected individuals (epistasis hypothesis) with observed individuals. Phenotypes Xo H Xa (Xo – Xa)2 : Xa c 2 Red 192 12/16 189, 75 5, 06 189, 75 0, 03 Orange 47 3/16 47, 43 0, 18 47, 43 0, 01 White 14 1/16 15, 81 3, 27 15, 81 0, 21 Total 253 16/16 253 Degrees of freedom = 3 -1 = 2 c 2 = 0, 25 P =80 - 90% Conclusion: Hypothesis accepted 0, 25


FILE 8: POINT MUTATIONS

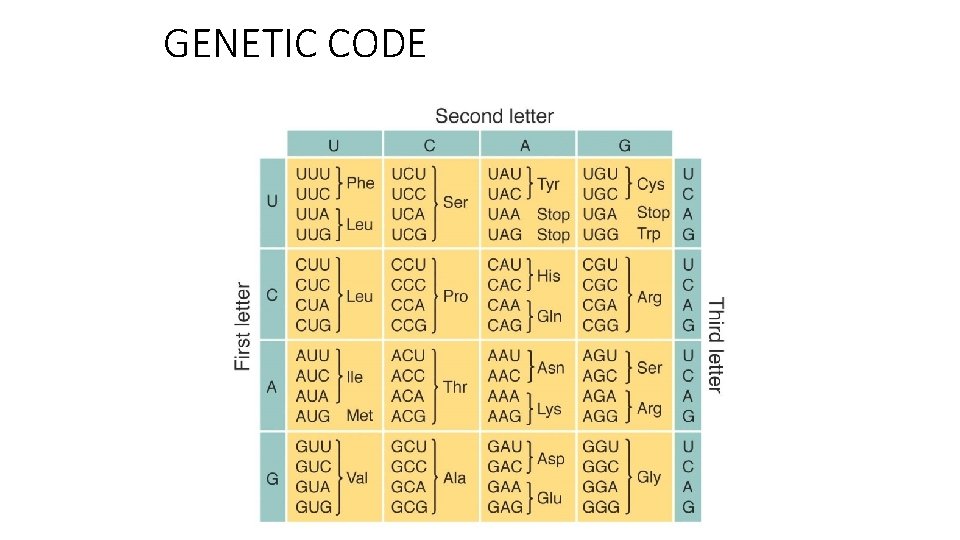
GENETIC CODE

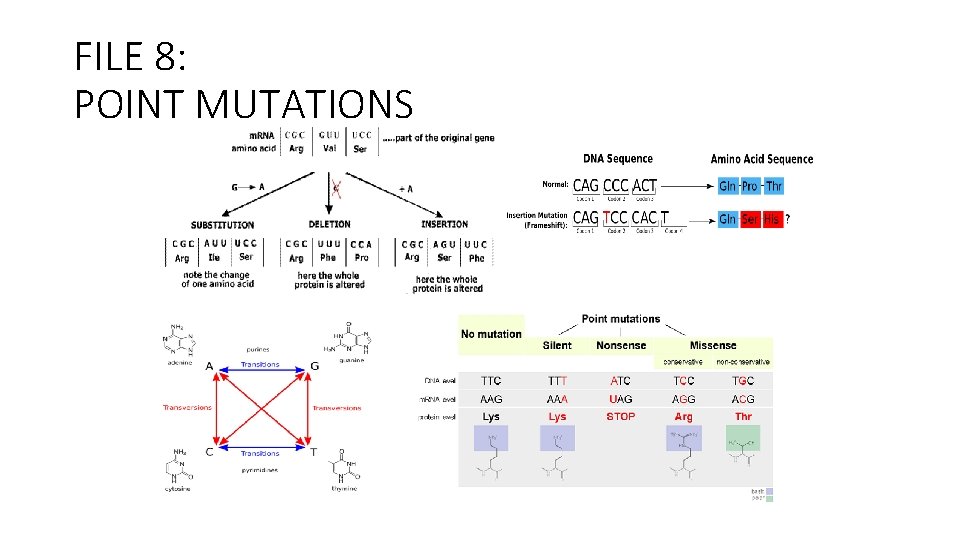
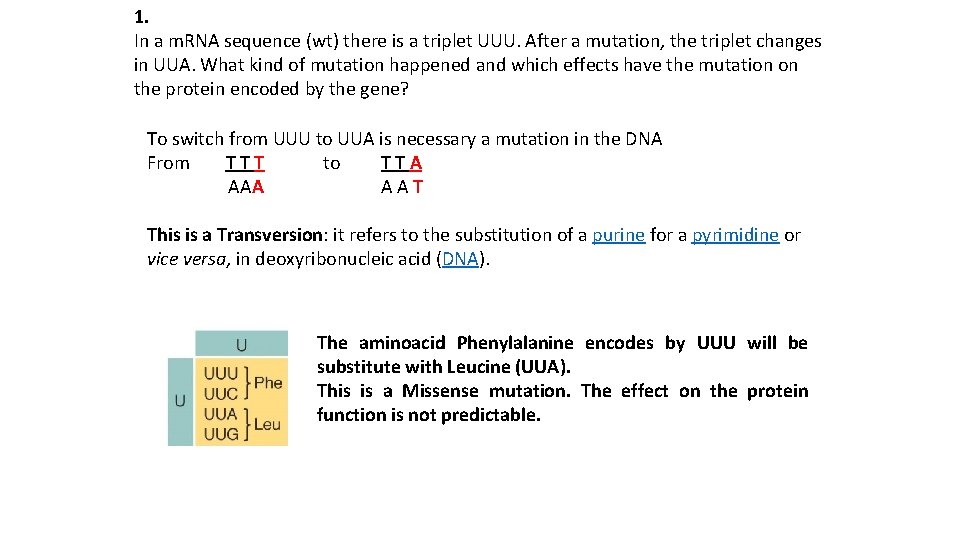
1. In a m. RNA sequence (wt) there is a triplet UUU. After a mutation, the triplet changes in UUA. What kind of mutation happened and which effects have the mutation on the protein encoded by the gene? To switch from UUU to UUA is necessary a mutation in the DNA From T T T to T T A AAA A A T This is a Transversion: it refers to the substitution of a purine for a pyrimidine or vice versa, in deoxyribonucleic acid (DNA). The aminoacid Phenylalanine encodes by UUU will be substitute with Leucine (UUA). This is a Missense mutation. The effect on the protein function is not predictable.

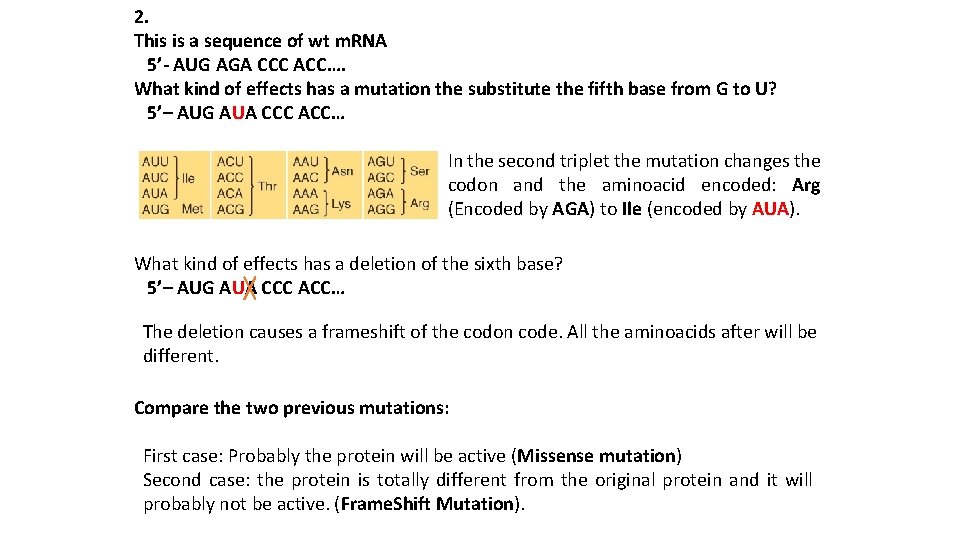
2. This is a sequence of wt m. RNA 5’- AUG AGA CCC ACC…. What kind of effects has a mutation the substitute the fifth base from G to U? 5’– AUG AUA CCC ACC… In the second triplet the mutation changes the codon and the aminoacid encoded: Arg (Encoded by AGA) to Ile (encoded by AUA). What kind of effects has a deletion of the sixth base? 5’– AUG AUA CCC ACC… The deletion causes a frameshift of the codon code. All the aminoacids after will be different. Compare the two previous mutations: First case: Probably the protein will be active (Missense mutation) Second case: the protein is totally different from the original protein and it will probably not be active. (Frame. Shift Mutation).

3. A nucleotide sequence below 5' 3' 5 10 15 20 25 30 35 ATTCGATGGCAGTGCCAAAGTGGTGATGGC TAAGCTACCGTCACGGTTTCACCACTACCG 3' 5' a) Knowing that the transcription of this sequence is from left to right (5’->3’), write the resulting m. RNA sequence: 5’ AUUCGAUGGCAGUGCCAAAGUGGUGAUGGC b) Knowing that this m. RNA sequence contains the translation start codon, identify the starting codon and indicate the amino acid sequence of the resulting peptide. Met- Gly- Trp-Gln-Cys- Gln- Ser-Gly- Asp- Gly

5' 3' ATTCGATGGCAGTGCCAAAGTGGTGATGGC TAAGCTACCGTCACG GTTTCACCACTACCG 3' 5' C) Find which consequences will have on the amino acid sequence: - a transition of the T base pair in position 18; In the DNA sequence: switch from TA to CG; In the m. RNA sequence there is a switch between U and C thus between the triplet UGC (Cys) to CGC (Arg) that causes a MIS-SENSE mutation.

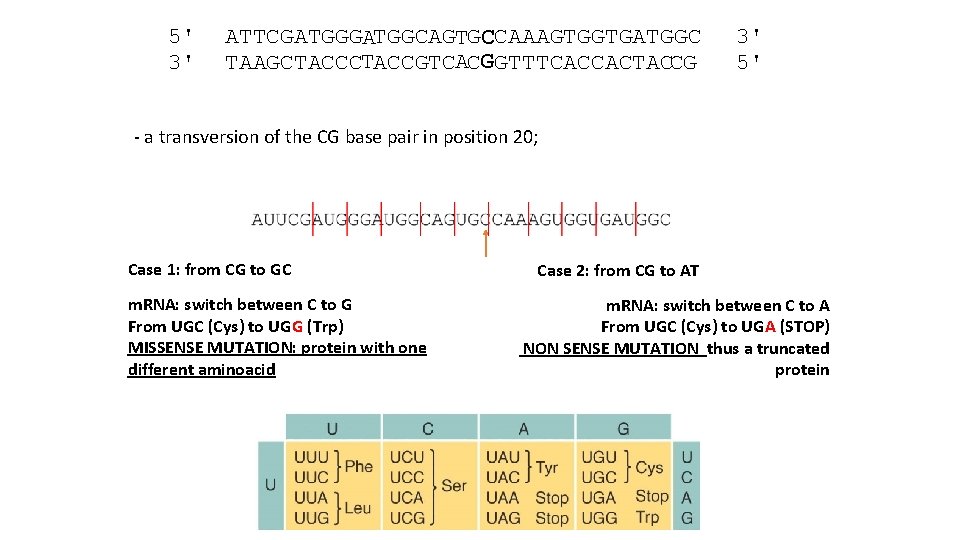
5' 3' ATTCGATGGCAGTGCCAAAGTGGTGATGGC TAAGCTACCGTCACG GTTTCACCACTACCG 3' 5' - a transversion of the CG base pair in position 20; Case 1: from CG to GC m. RNA: switch between C to G From UGC (Cys) to UGG (Trp) MISSENSE MUTATION: protein with one different aminoacid Case 2: from CG to AT m. RNA: switch between C to A From UGC (Cys) to UGA (STOP) NON SENSE MUTATION thus a truncated protein

5' 3' ATTCGATGGCAGTGCCAAAGTGGTGATGGC TAAGCTACCGTCACGGTTTCACCACTACCG 3' 5' - an insertion of a base pair after pair 11 (AT) +1 AUG GGA NUG GCA GUG CCA AAG UGA UGG C Met-Gly-aa. X-Ala-Val-Pro-Lys-Trp-Stop After the insertion all the aminoacids will be different: frame-shift mutation. The frame-shift creates a stop codon.

d) How can we abolish the insertion mutation in position 11? If in a position near the first insertion a deletion happens, we can restore the correct frame of aminoacids. For example in position 15 (intragenic suppressionsuppressor mutation) +1 AUG-GGA-NUG-GAG-UGC-CAA-AGU-GAU-GGC Met-Gly-aa. X-Glu-Cys-Gln-Ser-Gly-Asp-Gly The second mutation restore the correct reading frame. Only the aminoacids located between the two mutations will be different. DNA with insertion and subsequent suppressor mutation: 5’ ATTCGATGGGANTGGAGTGCCAAAGTGGTGATGGC 3’ 3’ TAAGCTACCCTNACCTCACGGTTTCACCACTACCG 5’

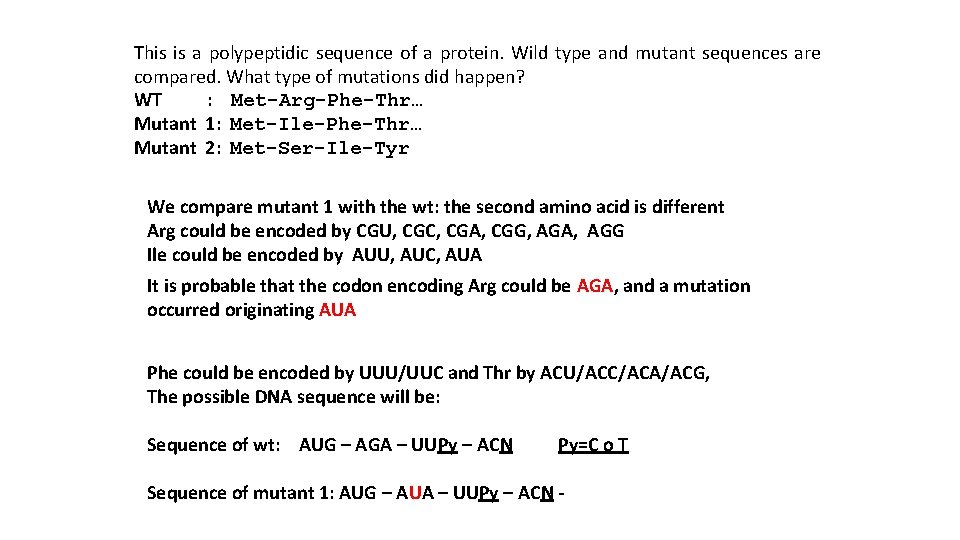
This is a polypeptidic sequence of a protein. Wild type and mutant sequences are compared. What type of mutations did happen? WT : Met-Arg-Phe-Thr… Mutant 1: Met-Ile-Phe-Thr… Mutant 2: Met-Ser-Ile-Tyr We compare mutant 1 with the wt: the second amino acid is different Arg could be encoded by CGU, CGC, CGA, CGG, AGA, AGG Ile could be encoded by AUU, AUC, AUA It is probable that the codon encoding Arg could be AGA, and a mutation occurred originating AUA Phe could be encoded by UUU/UUC and Thr by ACU/ACC/ACA/ACG, The possible DNA sequence will be: Sequence of wt: AUG – AGA – UUPy – ACN Py=C o T Sequence of mutant 1: AUG – AUA – UUPy – ACN -

WT : Met-Arg-Phe-Thr… Mutant 2: Met-Ser-Ile-Tyr We compare mutant 2 with wt: all the amicoacids after the Methionine are different. A frameshift mutation caused by an insertion. AUG – AGA – UUPy – ACN – Ser is encoded by UCN or AGU-AGC , We can hypotesize the sequence: AUG –AGN–AUU-Py. ACWith N=U/C The third codon AUU = Ile The fourth codon will be UAC = Tyr

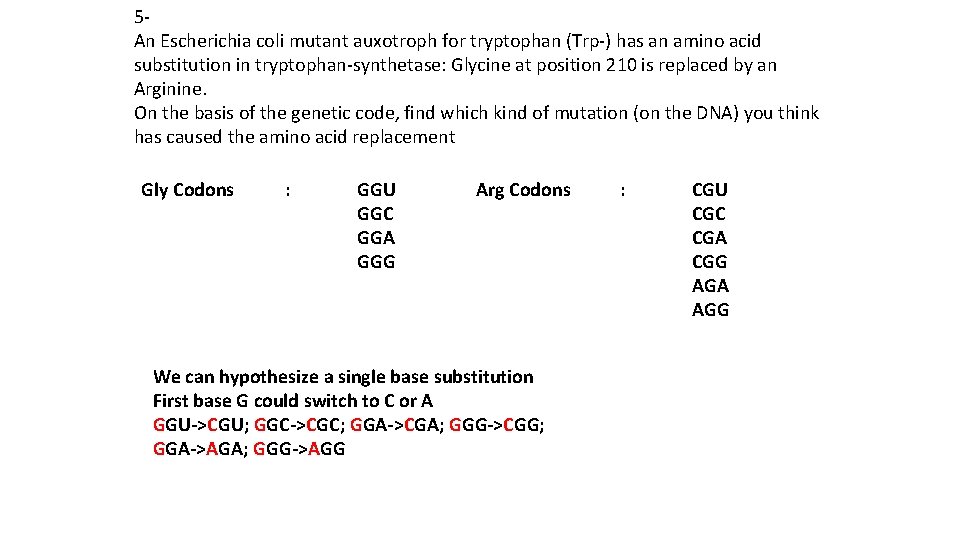
5 An Escherichia coli mutant auxotroph for tryptophan (Trp-) has an amino acid substitution in tryptophan-synthetase: Glycine at position 210 is replaced by an Arginine. On the basis of the genetic code, find which kind of mutation (on the DNA) you think has caused the amino acid replacement Gly Codons : GGU GGC GGA GGG Arg Codons We can hypothesize a single base substitution First base G could switch to C or A GGU->CGU; GGC->CGC; GGA->CGA; GGG->CGG; GGA->AGA; GGG->AGG : CGU CGC CGA CGG AGA AGG

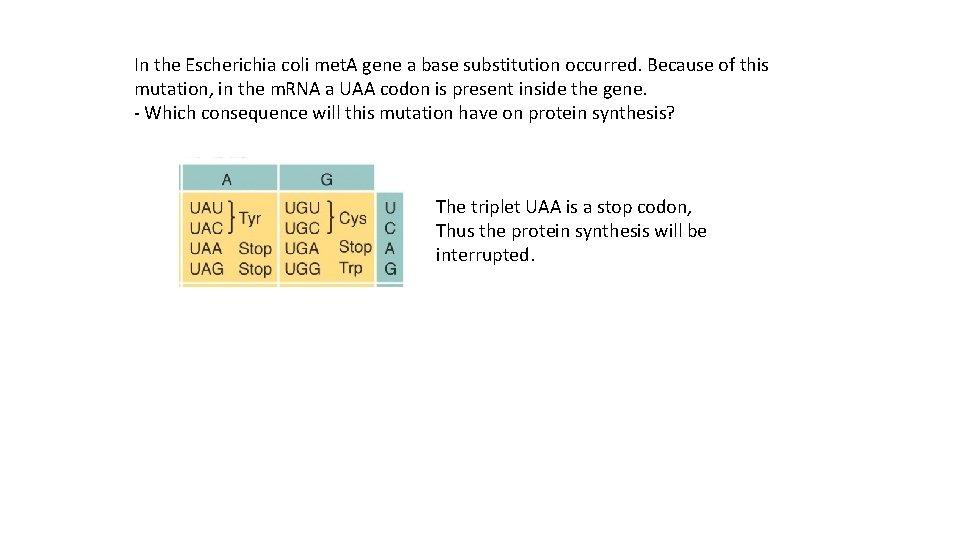
In the Escherichia coli met. A gene a base substitution occurred. Because of this mutation, in the m. RNA a UAA codon is present inside the gene. - Which consequence will this mutation have on protein synthesis? The triplet UAA is a stop codon, Thus the protein synthesis will be interrupted.

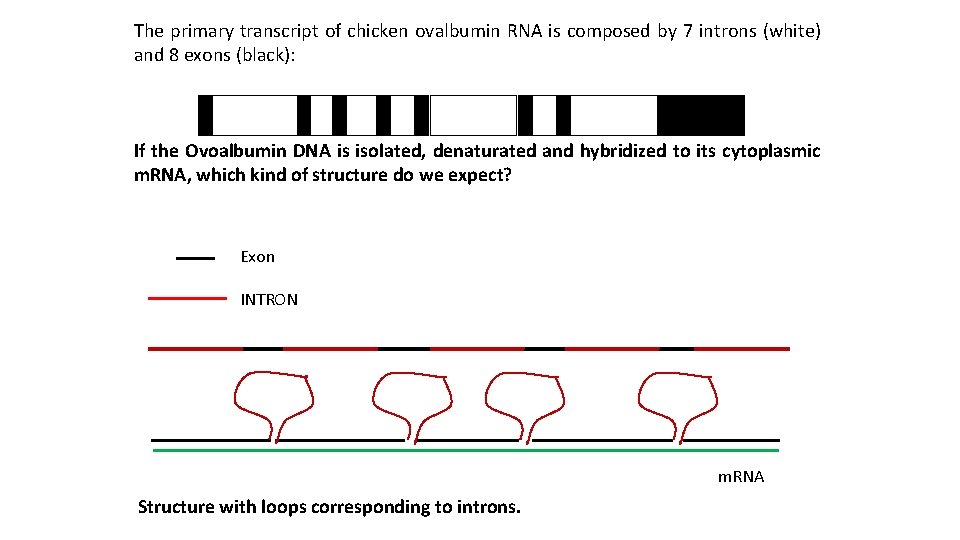
The primary transcript of chicken ovalbumin RNA is composed by 7 introns (white) and 8 exons (black): If the Ovoalbumin DNA is isolated, denaturated and hybridized to its cytoplasmic m. RNA, which kind of structure do we expect? Exon INTRON m. RNA Structure with loops corresponding to introns.

if a deletion of a base pair occurs in the middle of the second intron, which will be the likely effects on the resulting polypeptide? We don’t have any effect if the mutation is not in the splicing site. if a deletion of a base pair occurs in the middle of the first exon, which will be the likely effects on the resulting polypeptide? We have a frame-shift effect or a truncated protein.

FILE 8

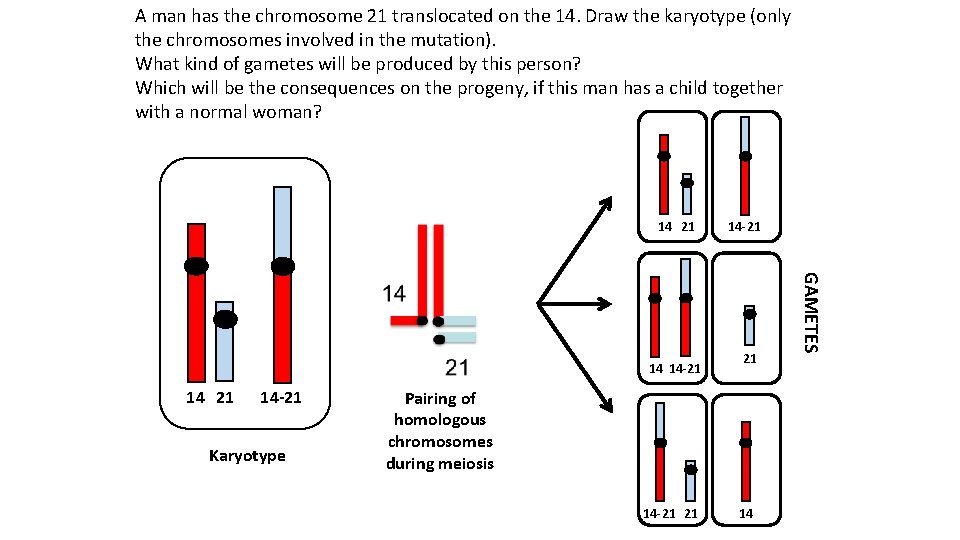
A man has the chromosome 21 translocated on the 14. Draw the karyotype (only the chromosomes involved in the mutation). What kind of gametes will be produced by this person? Which will be the consequences on the progeny, if this man has a child together with a normal woman? 14 21 14 -21 Karyotype 21 Pairing of homologous chromosomes during meiosis 14 -21 21 14 GAMETES 14 14 -21

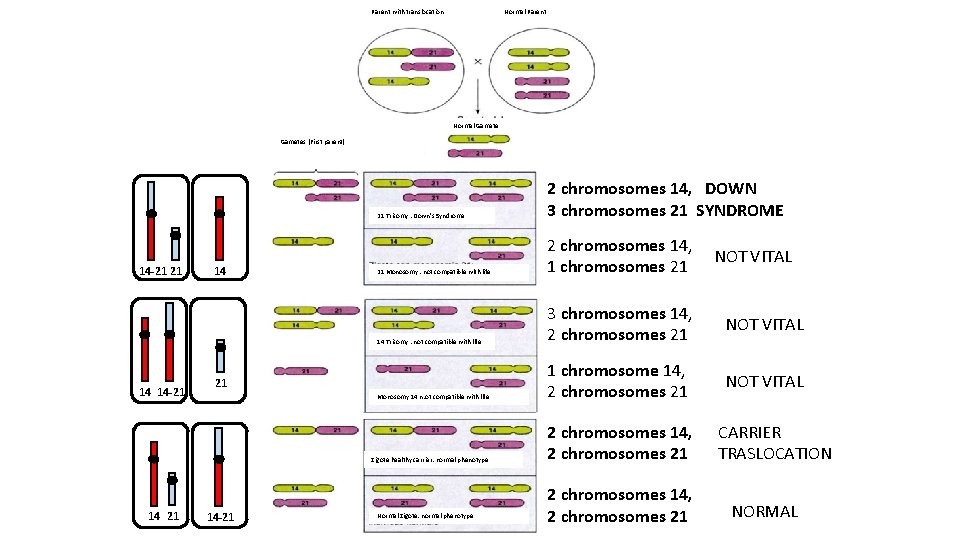
Parent with translocation Normal Parent Normal Gamete Gametes (First parent) 14 -21 21 14 14 -21 14 21 Trisomy : Down’s Syndrome 2 chromosomes 14, DOWN 3 chromosomes 21 SYNDROME 21 Monosomy : not compatible with life 2 chromosomes 14, NOT VITAL 1 chromosomes 21 14 Trisomy : not compatible with life 3 chromosomes 14, NOT VITAL 2 chromosomes 21 Monosomy 14 n: ot compatible with life 1 chromosome 14, NOT VITAL 2 chromosomes 21 21 Zigote healthy carrier: normal phenotype 14 21 14 -21 Normal Zigote: normal phenotype 2 chromosomes 14, 2 chromosomes 21 CARRIER TRASLOCATION NORMAL

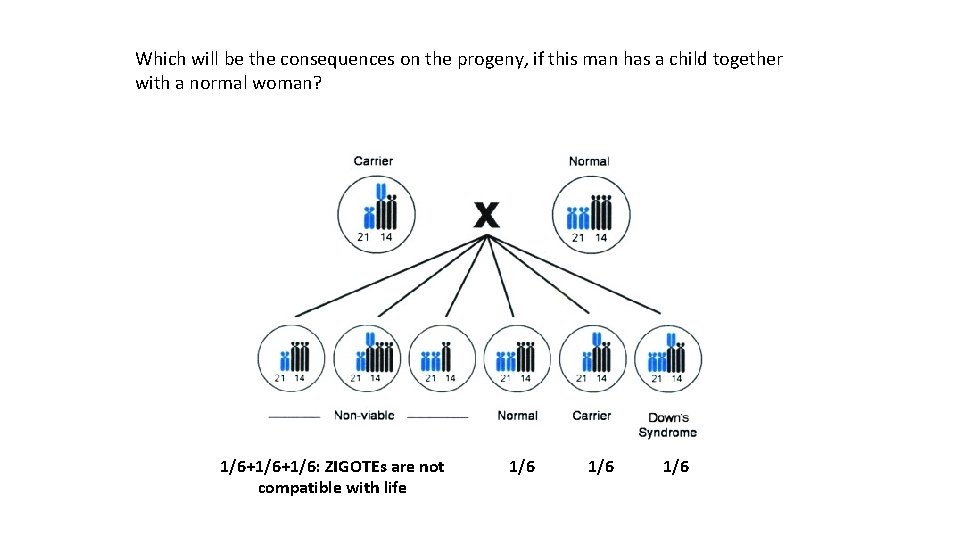
Which will be the consequences on the progeny, if this man has a child together with a normal woman? 1/6+1/6: ZIGOTEs are not compatible with life 1/6 1/6

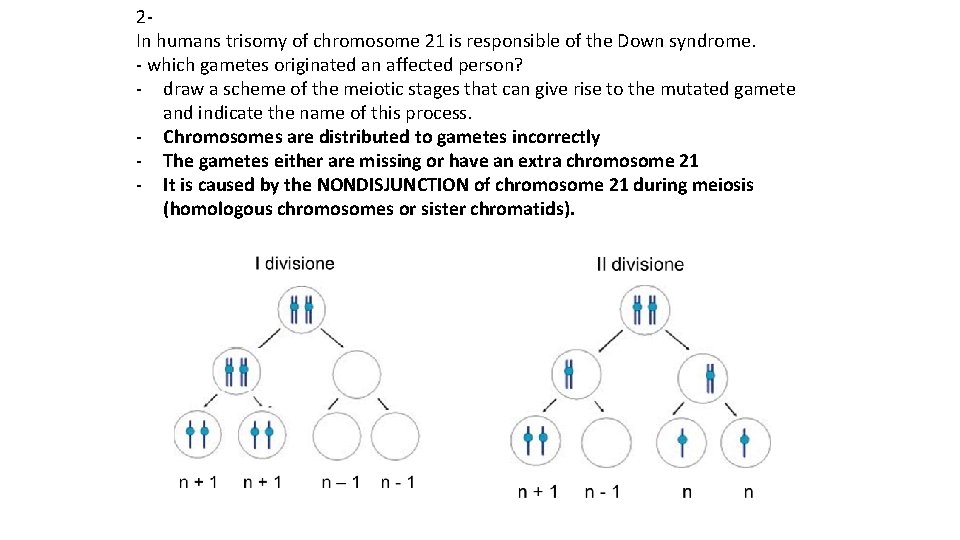
2 In humans trisomy of chromosome 21 is responsible of the Down syndrome. - which gametes originated an affected person? - draw a scheme of the meiotic stages that can give rise to the mutated gamete and indicate the name of this process. - Chromosomes are distributed to gametes incorrectly - The gametes either are missing or have an extra chromosome 21 - It is caused by the NONDISJUNCTION of chromosome 21 during meiosis (homologous chromosomes or sister chromatids).

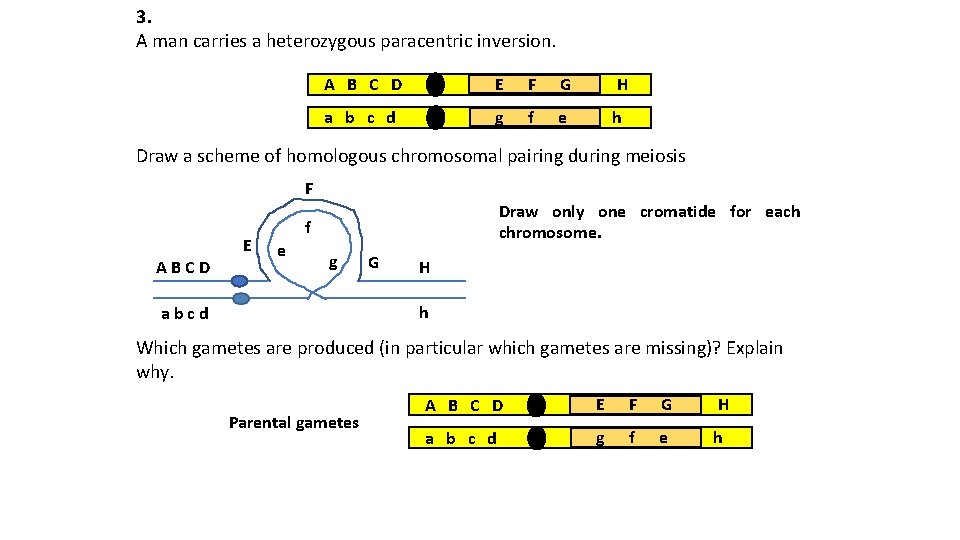
3. A man carries a heterozygous paracentric inversion. A B C D E F G H a b c d g f e h Draw a scheme of homologous chromosomal pairing during meiosis F ABCD E Draw only one cromatide for each chromosome. f e g G H h abcd Which gametes are produced (in particular which gametes are missing)? Explain why. Parental gametes A B C D E F G H a b c d g f e h

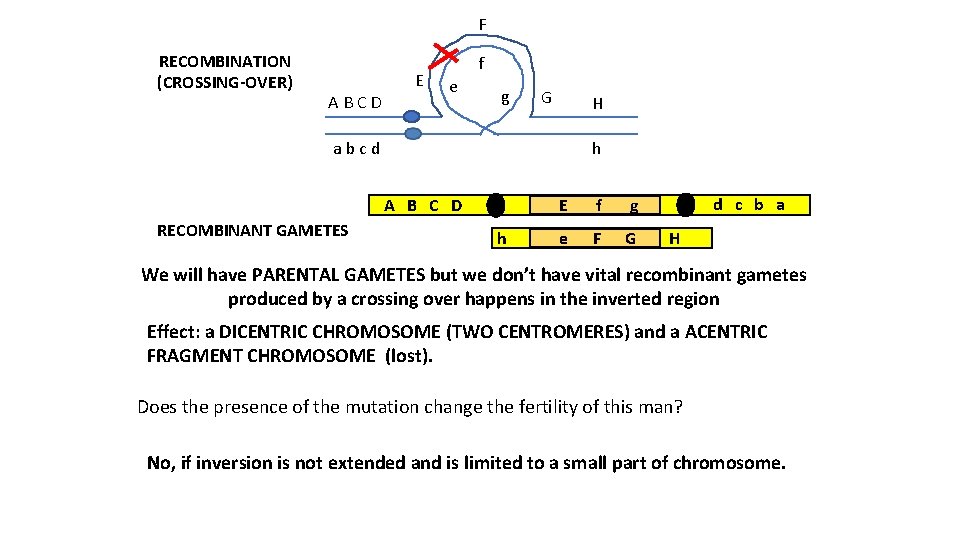
F RECOMBINATION (CROSSING-OVER) A B C D E f e g G H h a b c d A B C D RECOMBINANT GAMETES h E f g e F G d c b a H We will have PARENTAL GAMETES but we don’t have vital recombinant gametes produced by a crossing over happens in the inverted region Effect: a DICENTRIC CHROMOSOME (TWO CENTROMERES) and a ACENTRIC FRAGMENT CHROMOSOME (lost). Does the presence of the mutation change the fertility of this man? No, if inversion is not extended and is limited to a small part of chromosome.

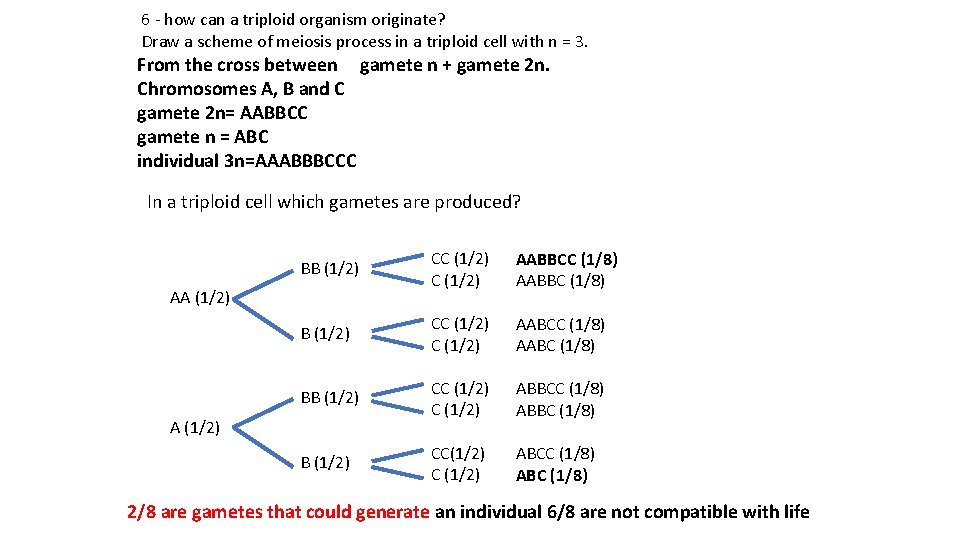
6 - how can a triploid organism originate? Draw a scheme of meiosis process in a triploid cell with n = 3. From the cross between gamete n + gamete 2 n. Chromosomes A, B and C gamete 2 n= AABBCC gamete n = ABC individual 3 n=AAABBBCCC In a triploid cell which gametes are produced? BB (1/2) CC (1/2) AABBCC (1/8) AABBC (1/8) B (1/2) CC (1/2) AABCC (1/8) AABC (1/8) BB (1/2) CC (1/2) ABBCC (1/8) ABBC (1/8) B (1/2) CC(1/2) C (1/2) ABCC (1/8) ABC (1/8) AA (1/2) 2/8 are gametes that could generate an individual 6/8 are not compatible with life

MUTANTS IN MICROORGANISMS

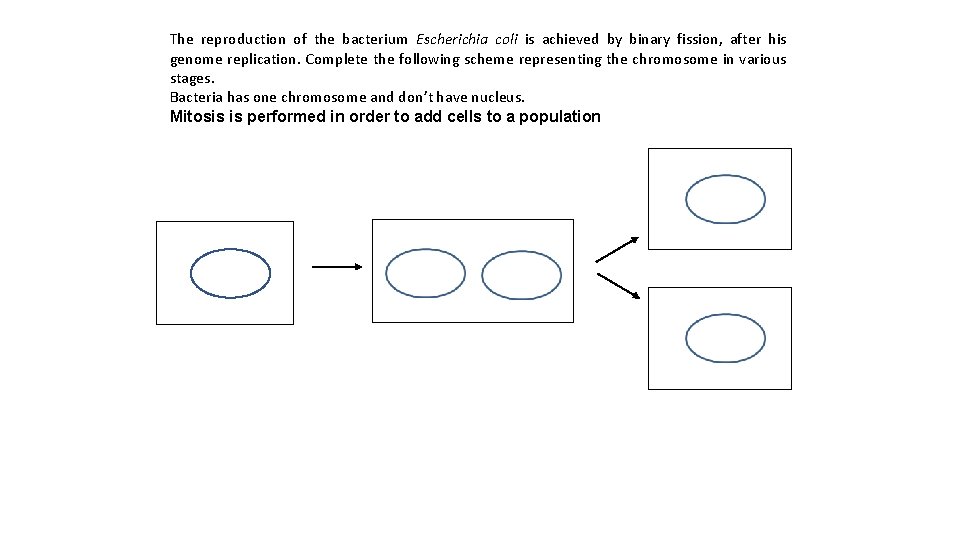
The reproduction of the bacterium Escherichia coli is achieved by binary fission, after his genome replication. Complete the following scheme representing the chromosome in various stages. Bacteria has one chromosome and don’t have nucleus. Mitosis is performed in order to add cells to a population

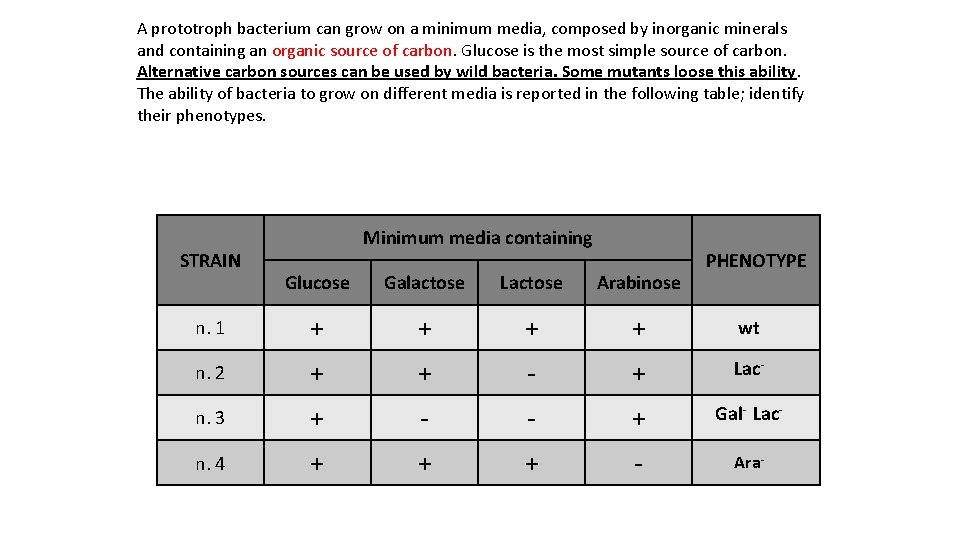
A prototroph bacterium can grow on a minimum media, composed by inorganic minerals and containing an organic source of carbon. Glucose is the most simple source of carbon. Alternative carbon sources can be used by wild bacteria. Some mutants loose this ability. The ability of bacteria to grow on different media is reported in the following table; identify their phenotypes. STRAIN Minimum media containing PHENOTYPE Glucose Galactose Lactose Arabinose n. 1 + + wt n. 2 + + - + Lac- n. 3 + - - + Gal- Lac- n. 4 + + + - Ara-

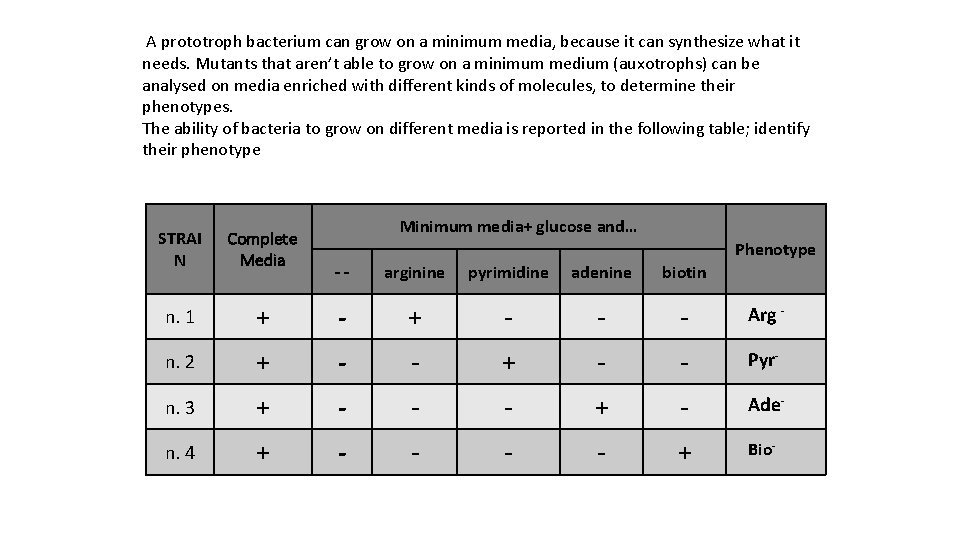
A prototroph bacterium can grow on a minimum media, because it can synthesize what it needs. Mutants that aren’t able to grow on a minimum medium (auxotrophs) can be analysed on media enriched with different kinds of molecules, to determine their phenotypes. The ability of bacteria to grow on different media is reported in the following table; identify their phenotype STRAI N Complete Media n. 1 Minimum media+ glucose and… Phenotype -- arginine pyrimidine adenine biotin + - - - Arg - n. 2 + - - Pyr- n. 3 + - - - + - Ade- n. 4 + - - + Bio-

A mutant bacterium isn’t able to synthesize the prolin amino acid and to metabolize lactose. Indicate which molecules must be added to a minimum media to grow this mutant. Minimum media + glucose and proline An Escherichia coli mutant carries a temperature-sensitive mutation in the polymerase III gene that is necessary for DNA replication. At which temperature can this bacterium grow? 30°C it grows 37°C it grows (optimal) 42°C it does not grow

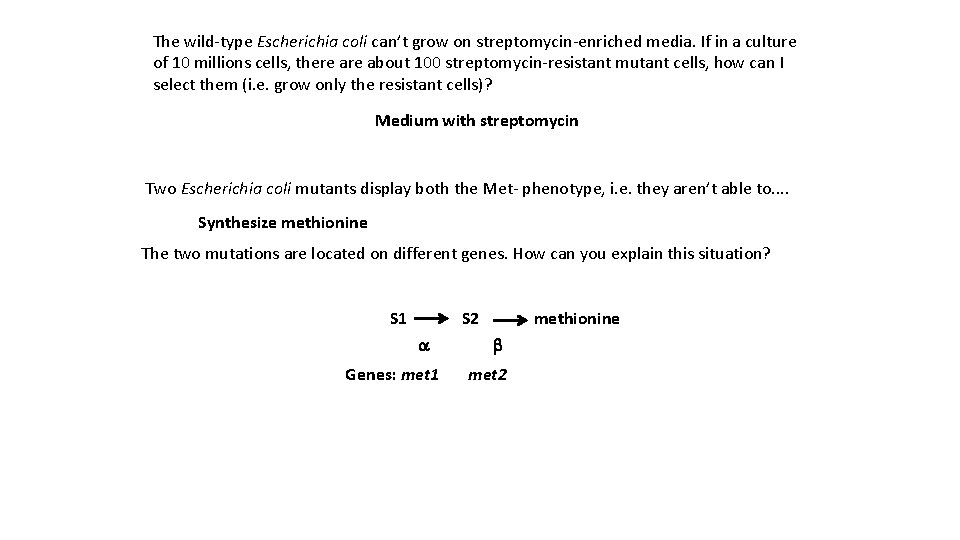
The wild-type Escherichia coli can’t grow on streptomycin-enriched media. If in a culture of 10 millions cells, there about 100 streptomycin-resistant mutant cells, how can I select them (i. e. grow only the resistant cells)? Medium with streptomycin Two Escherichia coli mutants display both the Met- phenotype, i. e. they aren’t able to. . Synthesize methionine The two mutations are located on different genes. How can you explain this situation? S 1 S 2 a Genes: met 1 methionine b met 2

Two Escherichia coli mutants can’t grow on galactose medium if galactose is the unique source of Carbon. - Gal • Assign the phenotypes at the mutants ……………. . • Are you sure that the mutations regard the same gene? No because the pathways of galactose degradation is composed by different genes. If only one of the genes is inactive the galactose is not metabolized.

FILE 10

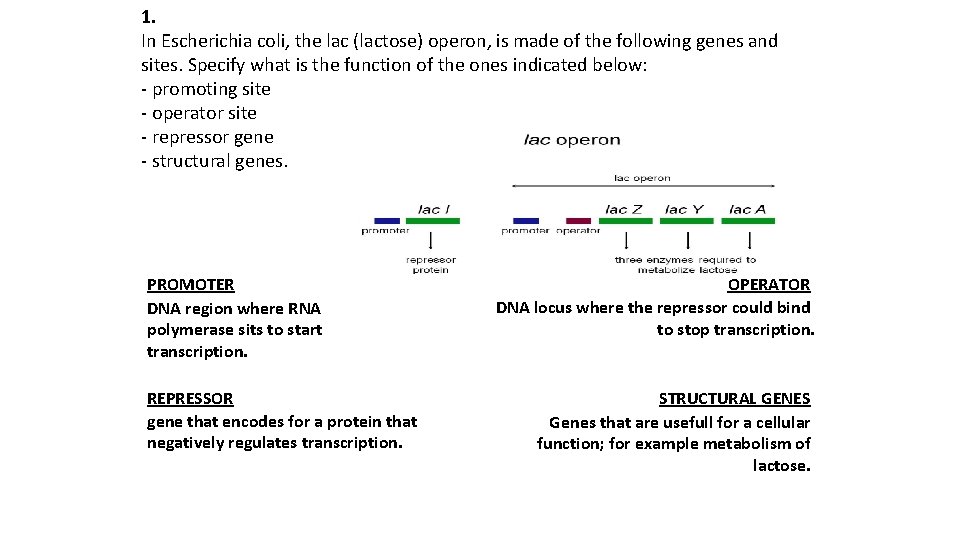
1. In Escherichia coli, the lac (lactose) operon, is made of the following genes and sites. Specify what is the function of the ones indicated below: - promoting site - operator site - repressor gene - structural genes. PROMOTER DNA region where RNA polymerase sits to start transcription. REPRESSOR gene that encodes for a protein that negatively regulates transcription. OPERATOR DNA locus where the repressor could bind to stop transcription. STRUCTURAL GENES Genes that are usefull for a cellular function; for example metabolism of lactose.

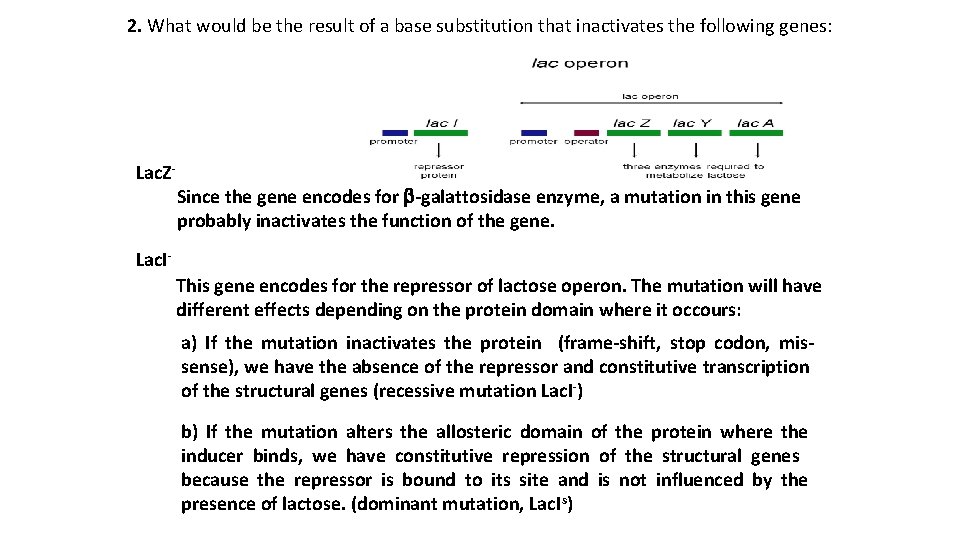
2. What would be the result of a base substitution that inactivates the following genes: Lac. Z- Since the gene encodes for b-galattosidase enzyme, a mutation in this gene probably inactivates the function of the gene. Lac. IThis gene encodes for the repressor of lactose operon. The mutation will have different effects depending on the protein domain where it occours: a) If the mutation inactivates the protein (frame-shift, stop codon, missense), we have the absence of the repressor and constitutive transcription of the structural genes (recessive mutation Lac. I-) b) If the mutation alters the allosteric domain of the protein where the inducer binds, we have constitutive repression of the structural genes because the repressor is bound to its site and is not influenced by the presence of lactose. (dominant mutation, Lac. Is)

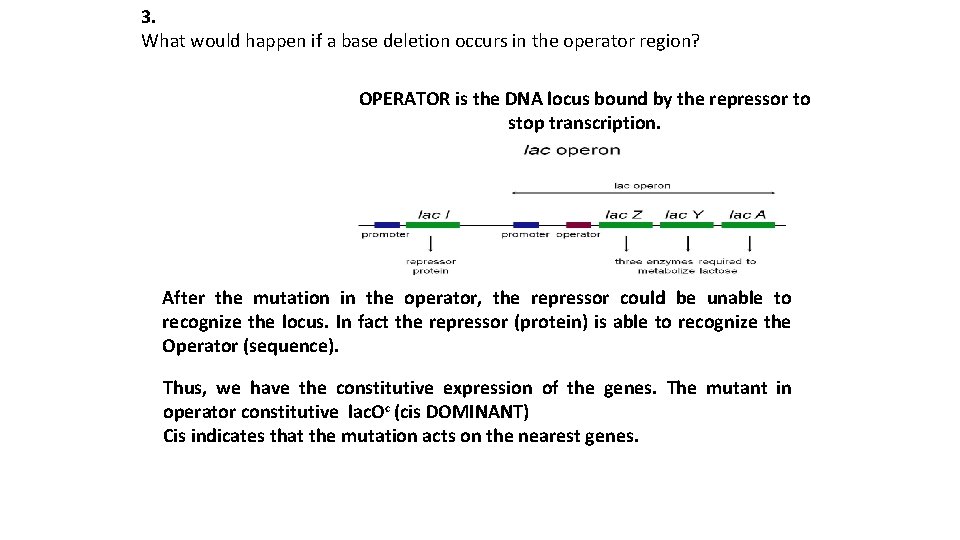
3. What would happen if a base deletion occurs in the operator region? OPERATOR is the DNA locus bound by the repressor to stop transcription. After the mutation in the operator, the repressor could be unable to recognize the locus. In fact the repressor (protein) is able to recognize the Operator (sequence). Thus, we have the constitutive expression of the genes. The mutant in operator constitutive lac. Oc (cis DOMINANT) Cis indicates that the mutation acts on the nearest genes.

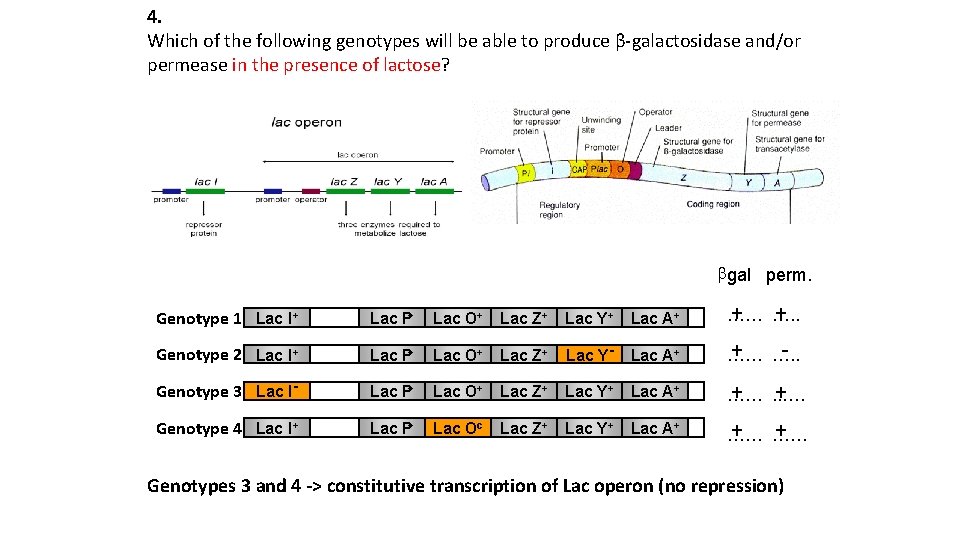
4. Which of the following genotypes will be able to produce β-galactosidase and/or permease in the presence of lactose? bgal perm. + + …… …. . Genotype 1 Lac I+ Lac P+ Lac O+ Lac Z+ Lac Y+ Lac A+ + … …. . Genotype 2 Lac I+ Lac P+ Lac O+ Lac Z+ Lac Y- Lac A+ … Lac P+ Lac O+ Lac Z+ Lac Y+ Lac A+ Genotype 3 Lac I+ + …… . . …. Genotype 4 Lac I+ Lac P+ Lac Oc Lac Z+ Lac Y+ Lac A+ + … …… + … Genotypes 3 and 4 -> constitutive transcription of Lac operon (no repression)

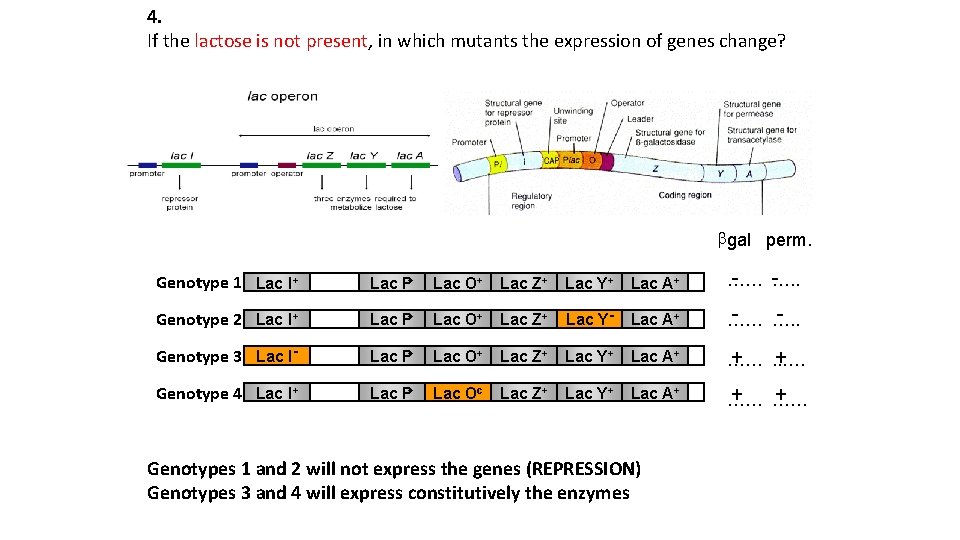
4. If the lactose is not present, in which mutants the expression of genes change? bgal perm. - …… …. . Genotype 1 Lac I+ Lac P+ Lac O+ Lac Z+ Lac Y+ Lac A+ - … …. . Genotype 2 Lac I+ Lac P+ Lac O+ Lac Z+ Lac Y- Lac A+ … Lac P+ Lac O+ Lac Z+ Lac Y+ Lac A+ Genotype 3 Lac I+ + …… . . …. Genotype 4 Lac I+ Lac P+ Lac Oc Lac Z+ Lac Y+ Lac A+ + … …… + … Genotypes 1 and 2 will not express the genes (REPRESSION) Genotypes 3 and 4 will express constitutively the enzymes

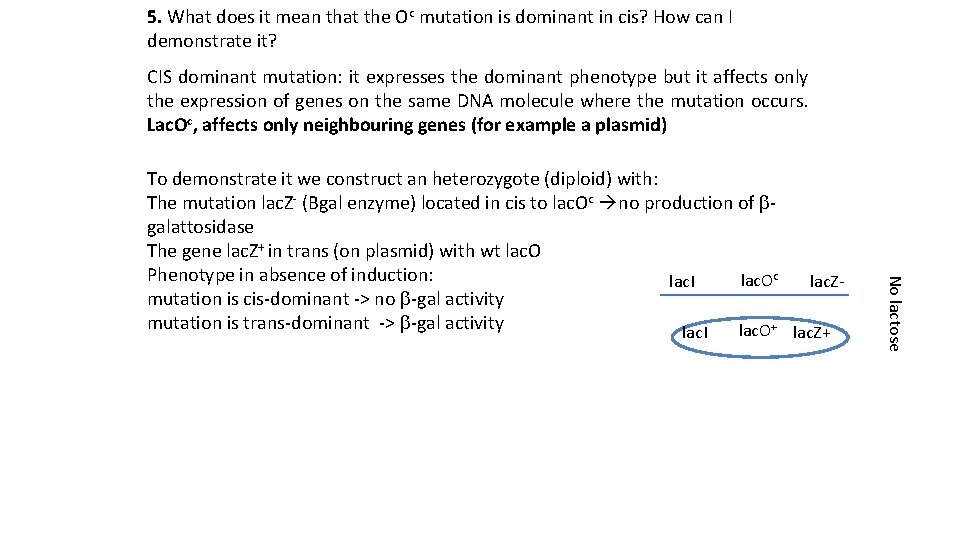
5. What does it mean that the Oc mutation is dominant in cis? How can I demonstrate it? CIS dominant mutation: it expresses the dominant phenotype but it affects only the expression of genes on the same DNA molecule where the mutation occurs. Lac. Oc, affects only neighbouring genes (for example a plasmid) No lactose To demonstrate it we construct an heterozygote (diploid) with: The mutation lac. Z- (Bgal enzyme) located in cis to lac. Oc no production of bgalattosidase The gene lac. Z+ in trans (on plasmid) with wt lac. O Phenotype in absence of induction: lac. Oc lac. I lac. Zmutation is cis-dominant -> no b-gal activity mutation is trans-dominant -> b-gal activity lac. O+ lac. Z+ lac. I

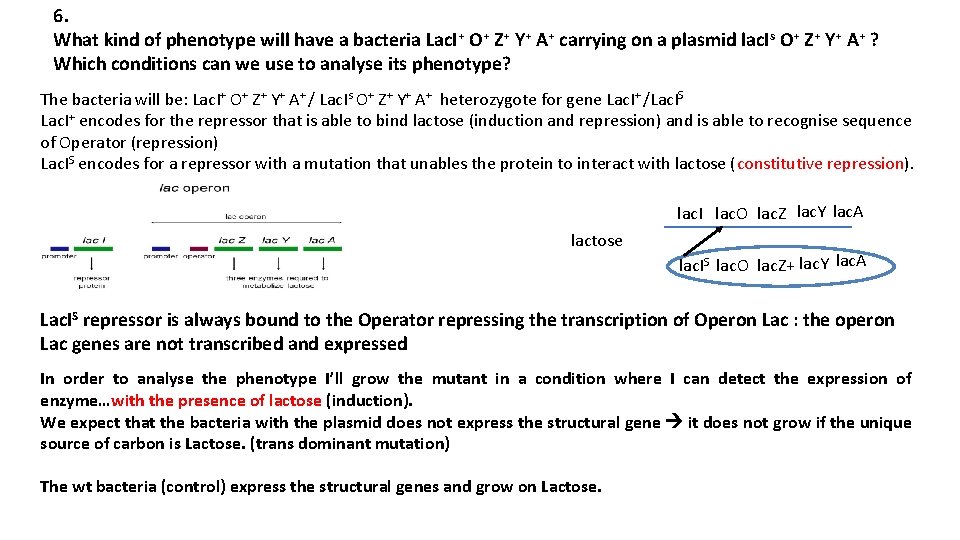
6. What kind of phenotype will have a bacteria Lac. I+ O+ Z+ Y+ A+ carrying on a plasmid lac. Is O+ Z+ Y+ A+ ? Which conditions can we use to analyse its phenotype? The bacteria will be: Lac. I+ O+ Z+ Y+ A+ / Lac. Is O+ Z+ Y+ A+ heterozygote for gene Lac. I+ /Lac. IS Lac. I+ encodes for the repressor that is able to bind lactose (induction and repression) and is able to recognise sequence of Operator (repression) Lac. IS encodes for a repressor with a mutation that unables the protein to interact with lactose (constitutive repression). lac. I lac. O lac. Z lac. Y lac. A lactose lac. IS lac. O lac. Z+ lac. Y lac. A Lac. IS repressor is always bound to the Operator repressing the transcription of Operon Lac : the operon Lac genes are not transcribed and expressed In order to analyse the phenotype I’ll grow the mutant in a condition where I can detect the expression of enzyme…with the presence of lactose (induction). We expect that the bacteria with the plasmid does not express the structural gene it does not grow if the unique source of carbon is Lactose. (trans dominant mutation) The wt bacteria (control) express the structural genes and grow on Lactose.

7. Two bacteria have a Trp- phenotype, then? The Trp- bacteria are unable to synthesize Tryptophan. It needs Tryptophan to grow. I have to add Tryptophan in the medium. How can I verify whether the two mutations are in the same gene? I complement them: I produce bacteria partially diploid carrying both mutations: one on a plasmid with all the operon Trp, the other on the chromosome. To analyse the phenotype I plate them on a medium without Tryptophan CASE 1: If bacteria grow, the two mutations complement each other, because they affect two different genes. CASE 2: If bacteria don’t grow, the two mutations do not complement each other, because they affect the same gene.

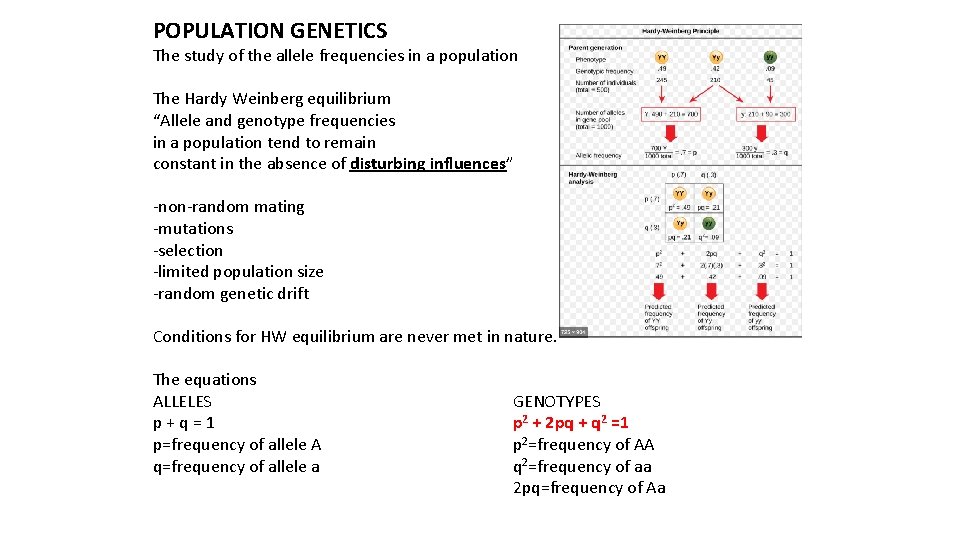
POPULATION GENETICS The study of the allele frequencies in a population The Hardy Weinberg equilibrium “Allele and genotype frequencies in a population tend to remain constant in the absence of disturbing influences” -non-random mating -mutations -selection -limited population size -random genetic drift Conditions for HW equilibrium are never met in nature. The equations ALLELES p + q = 1 p=frequency of allele A q=frequency of allele a GENOTYPES p 2 + 2 pq + q 2 =1 p 2=frequency of AA q 2=frequency of aa 2 pq=frequency of Aa

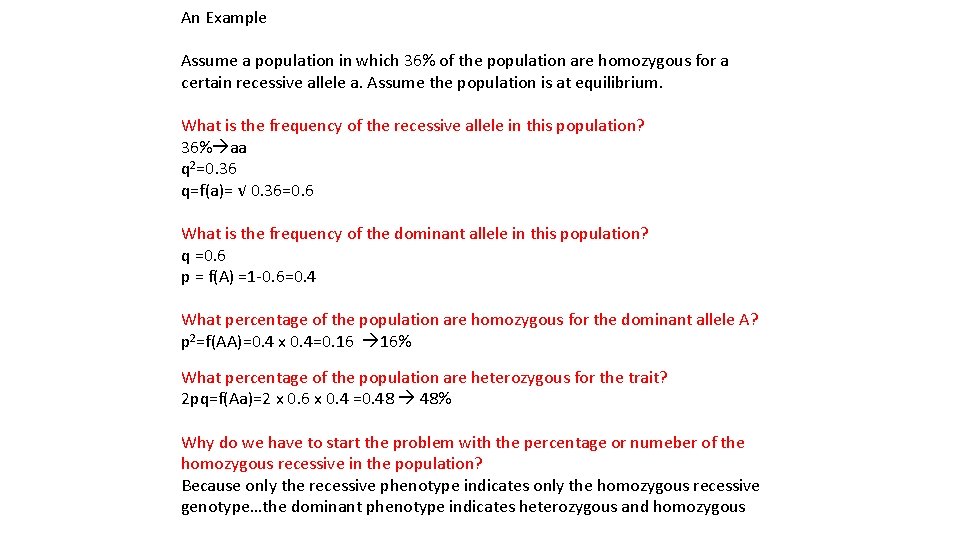
An Example Assume a population in which 36% of the population are homozygous for a certain recessive allele a. Assume the population is at equilibrium. What is the frequency of the recessive allele in this population? 36% aa q 2=0. 36 q=f(a)= √ 0. 36=0. 6 What is the frequency of the dominant allele in this population? q =0. 6 p = f(A) =1 -0. 6=0. 4 What percentage of the population are homozygous for the dominant allele A? p 2=f(AA)=0. 4 x 0. 4=0. 16 16% What percentage of the population are heterozygous for the trait? 2 pq=f(Aa)=2 x 0. 6 x 0. 4 =0. 48 48% Why do we have to start the problem with the percentage or numeber of the homozygous recessive in the population? Because only the recessive phenotype indicates only the homozygous recessive genotype…the dominant phenotype indicates heterozygous and homozygous

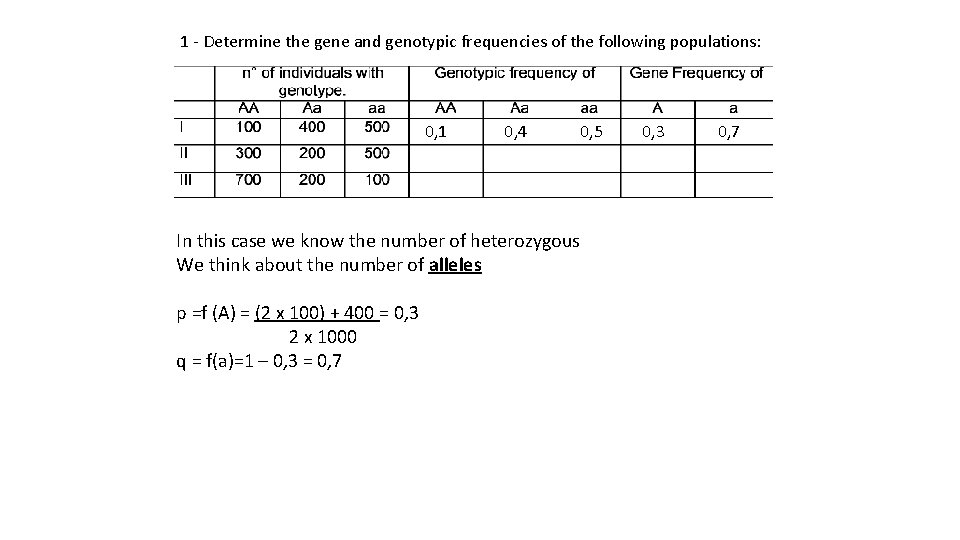
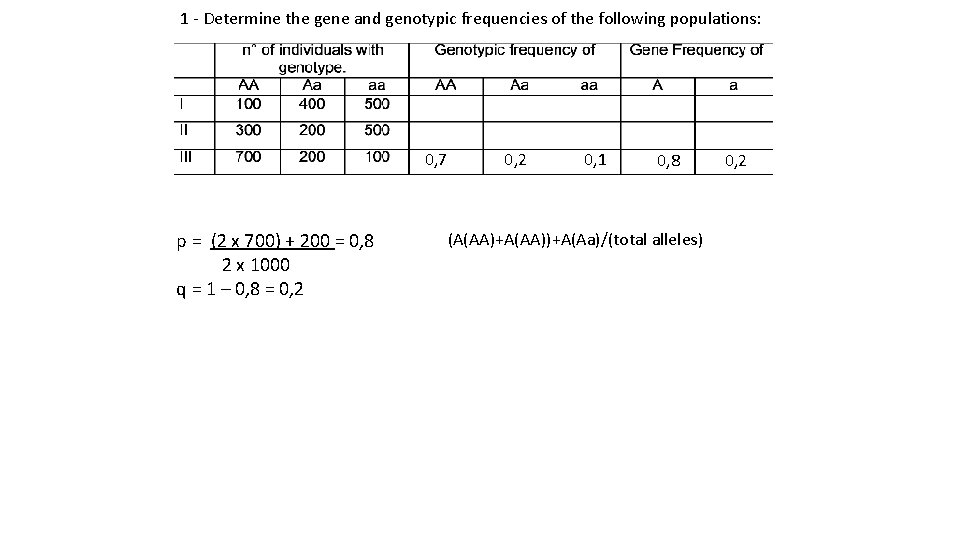
1 - Determine the gene and genotypic frequencies of the following populations: 0, 1 0, 4 0, 5 In this case we know the number of heterozygous We think about the number of alleles p =f (A) = (2 x 100) + 400 = 0, 3 2 x 1000 q = f(a)=1 – 0, 3 = 0, 7 0, 3 0, 7

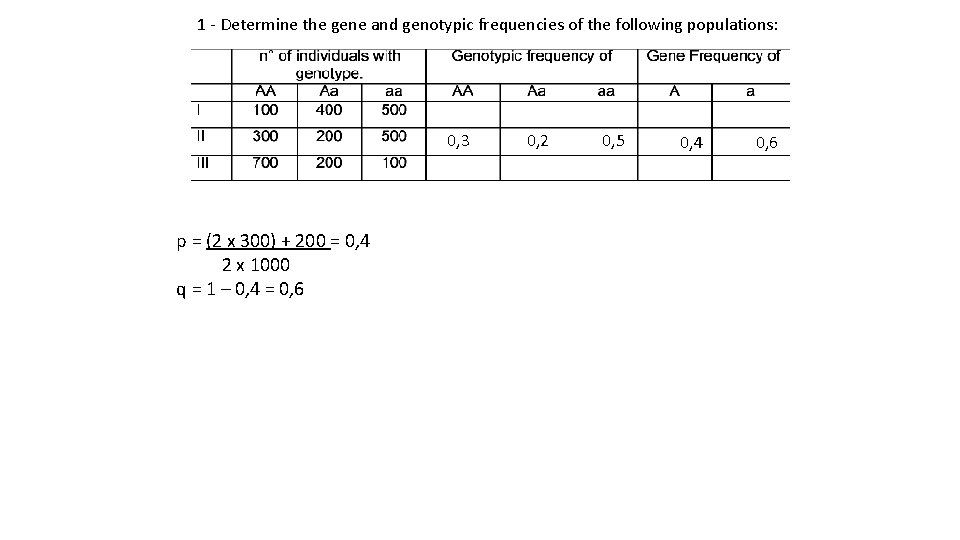
1 - Determine the gene and genotypic frequencies of the following populations: 0, 3 0, 2 0, 5 p = (2 x 300) + 200 = 0, 4 2 x 1000 q = 1 – 0, 4 = 0, 6 0, 4 0, 6

1 - Determine the gene and genotypic frequencies of the following populations: 0, 7 0, 2 0, 1 p = (2 x 700) + 200 = 0, 8 2 x 1000 q = 1 – 0, 8 = 0, 2 0, 8 0, 2 (A(AA)+A(AA))+A(Aa)/(total alleles)

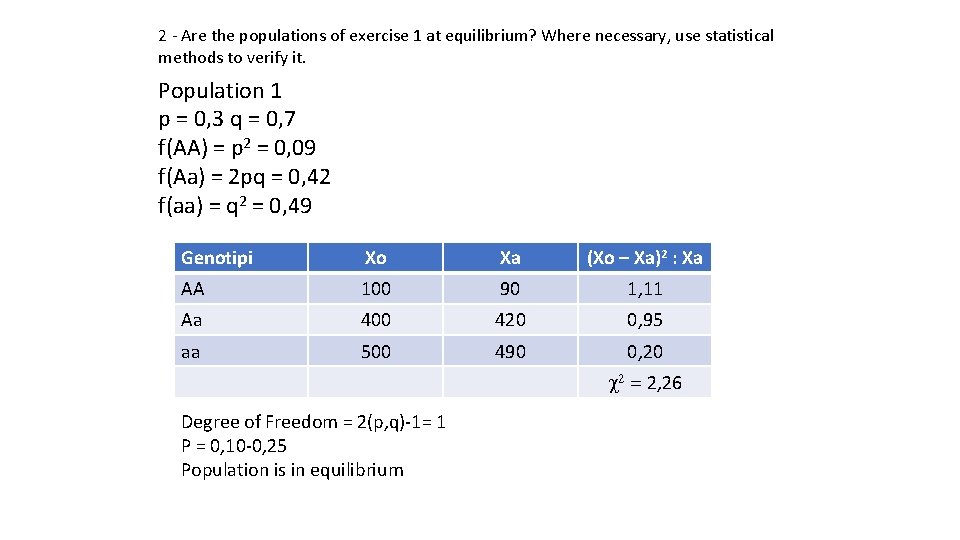
2 - Are the populations of exercise 1 at equilibrium? Where necessary, use statistical methods to verify it. Population 1 p = 0, 3 q = 0, 7 f(AA) = p 2 = 0, 09 f(Aa) = 2 pq = 0, 42 f(aa) = q 2 = 0, 49 Genotipi Xo Xa (Xo – Xa)2 : Xa AA 100 90 1, 11 Aa 400 420 0, 95 aa 500 490 0, 20 c 2 = 2, 26 Degree of Freedom = 2(p, q)-1= 1 P = 0, 10 -0, 25 Population is in equilibrium

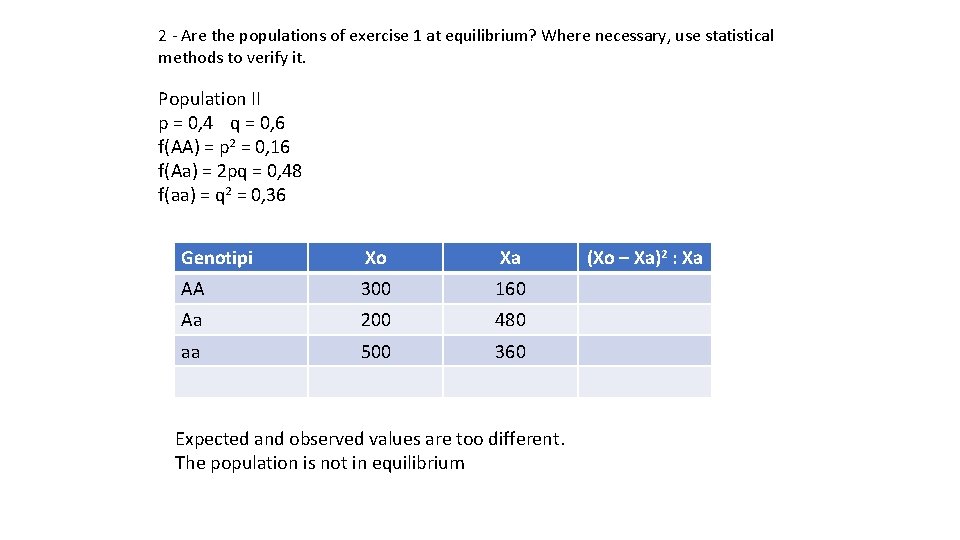
2 - Are the populations of exercise 1 at equilibrium? Where necessary, use statistical methods to verify it. Population II p = 0, 4 q = 0, 6 f(AA) = p 2 = 0, 16 f(Aa) = 2 pq = 0, 48 f(aa) = q 2 = 0, 36 Genotipi Xo Xa AA 300 160 Aa 200 480 aa 500 360 Expected and observed values are too different. The population is not in equilibrium (Xo – Xa)2 : Xa

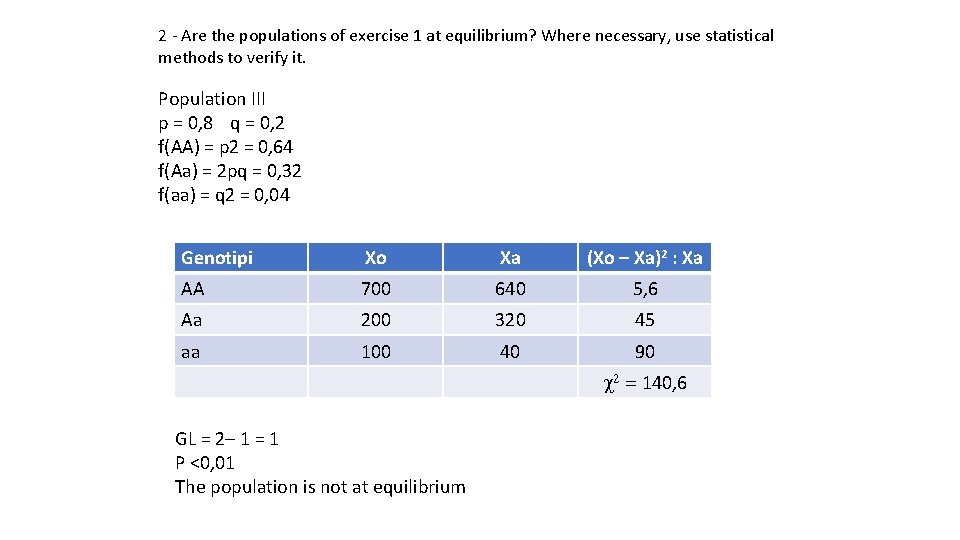
2 - Are the populations of exercise 1 at equilibrium? Where necessary, use statistical methods to verify it. Population III p = 0, 8 q = 0, 2 f(AA) = p 2 = 0, 64 f(Aa) = 2 pq = 0, 32 f(aa) = q 2 = 0, 04 Genotipi Xo Xa (Xo – Xa)2 : Xa AA 700 640 5, 6 Aa 200 320 45 aa 100 40 90 c 2 = 140, 6 GL = 2– 1 = 1 P <0, 01 The population is not at equilibrium

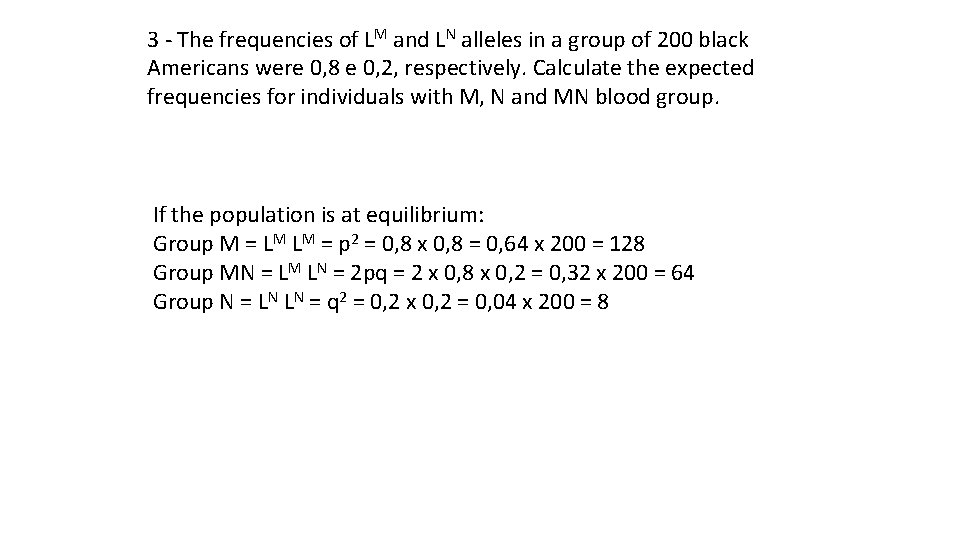
3 - The frequencies of LM and LN alleles in a group of 200 black Americans were 0, 8 e 0, 2, respectively. Calculate the expected frequencies for individuals with M, N and MN blood group. If the population is at equilibrium: Group M = LM LM = p 2 = 0, 8 x 0, 8 = 0, 64 x 200 = 128 Group MN = LM LN = 2 pq = 2 x 0, 8 x 0, 2 = 0, 32 x 200 = 64 Group N = LN LN = q 2 = 0, 2 x 0, 2 = 0, 04 x 200 = 8

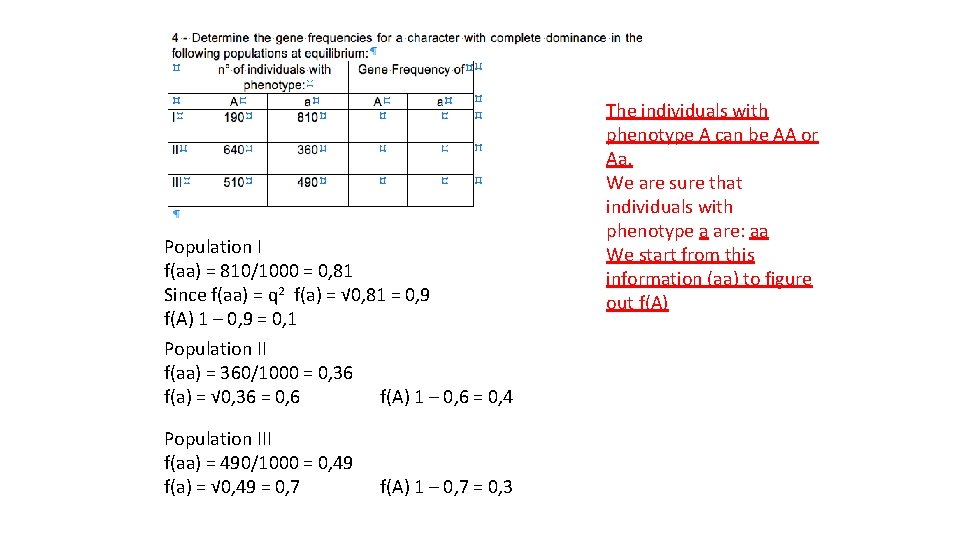
Population I f(aa) = 810/1000 = 0, 81 Since f(aa) = q 2 f(a) = √ 0, 81 = 0, 9 f(A) 1 – 0, 9 = 0, 1 Population II f(aa) = 360/1000 = 0, 36 f(a) = √ 0, 36 = 0, 6 f(A) 1 – 0, 6 = 0, 4 Population III f(aa) = 490/1000 = 0, 49 f(a) = √ 0, 49 = 0, 7 f(A) 1 – 0, 7 = 0, 3 The individuals with phenotype A can be AA or Aa. We are sure that individuals with phenotype a are: aa We start from this information (aa) to figure out f(A)

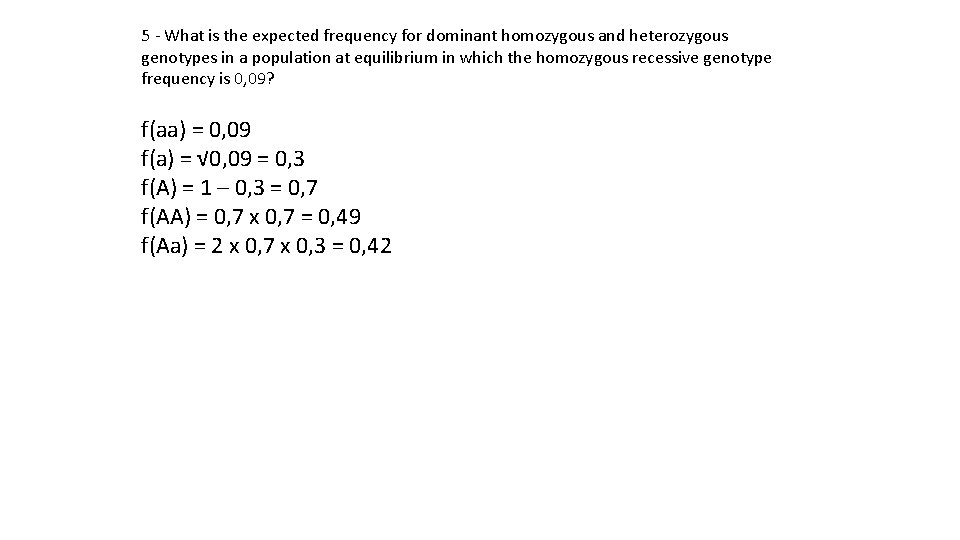
5 - What is the expected frequency for dominant homozygous and heterozygous genotypes in a population at equilibrium in which the homozygous recessive genotype frequency is 0, 09? f(aa) = 0, 09 f(a) = √ 0, 09 = 0, 3 f(A) = 1 – 0, 3 = 0, 7 f(AA) = 0, 7 x 0, 7 = 0, 49 f(Aa) = 2 x 0, 7 x 0, 3 = 0, 42

6 - if in a population at equilibrium the frequency of Rh- phenotype is 0, 0025, which is the expected frequency of heterozygous individuals? f(Rh-) = 0, 0025 = f(dd) f(d) = √ 0, 0025 = 0, 05 f(D) = 1 – 0, 05 = 0, 95 f(Dd) = 2 x 0, 95 x 0, 05 = 0, 095

7 In Drosophila melanogaster the w recessive sex-linked allele, is responsible of the white colour of the eyes. In a population the frequency of this allele is 0, 3. Which frequencies of male and female with white-eyes is expected if the population is at equilibrium? The frequency of these two alleles in the entire population are �� and ��. Let's assume the locus is present on the X chromosome. We will assume that the allele frequency do not differ between males and females. Male white eyes = w Y Female white eyes = w w Frequency: Males white eyes = f(w)=q = 0, 3 Females white eyes = f(ww) = q 2 = 0, 3 x 0, 3 = 0, 09 If alleles are X-linked, females may be homozygous or heterozygous, but males carry only a single allele for each X-linked locus. For X-linked alleles in females, the H-W frequencies are the same as those for autosomal loci. p 2+2 pq+q 2=1 In males, however, the frequencies of the genotypes will be p and q, the same as the frequencies of the alleles in the population. p+q=1

8 - Daltonism is due to a sex-linked recessive allele. In a population the daltonic male frequency is 0, 1; which is the one of daltonic females? Daltonic MALE = d Y (XY) hemizygote freq d. Y = freq allele d Daltonic FEMALE = d d (XX) Frequency Daltonic Males = 0, 1 Frequency allele d in MALES = 0, 1 Daltonic FEMALES = f(d d) = 0, 1 x 0, 1 = 0, 01 Healthy carriers= 2*0. 1*0. 9=0. 18

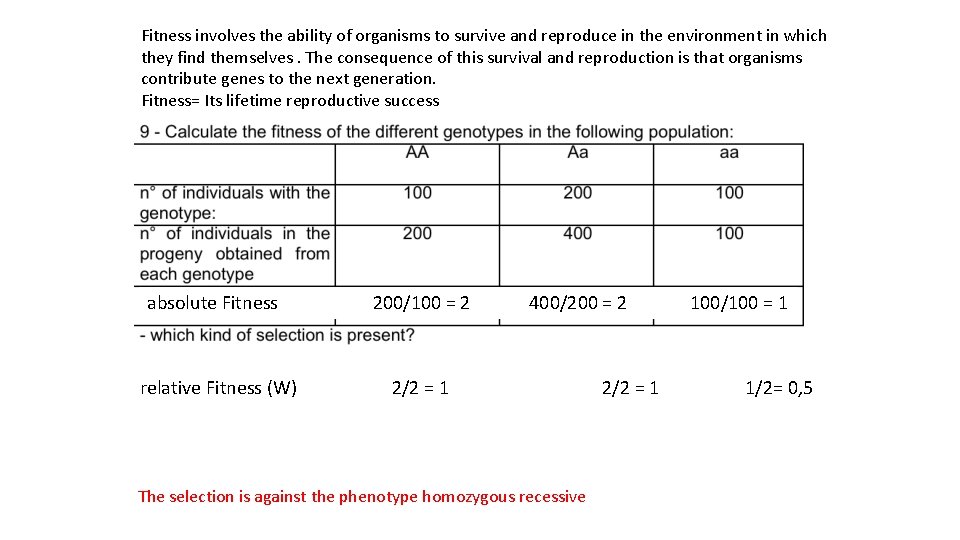
Fitness involves the ability of organisms to survive and reproduce in the environment in which they find themselves. The consequence of this survival and reproduction is that organisms contribute genes to the next generation. Fitness= Its lifetime reproductive success absolute Fitness 200/100 = 2 400/200 = 2 100/100 = 1 relative Fitness (W) 2/2 = 1 1/2= 0, 5 The selection is against the phenotype homozygous recessive

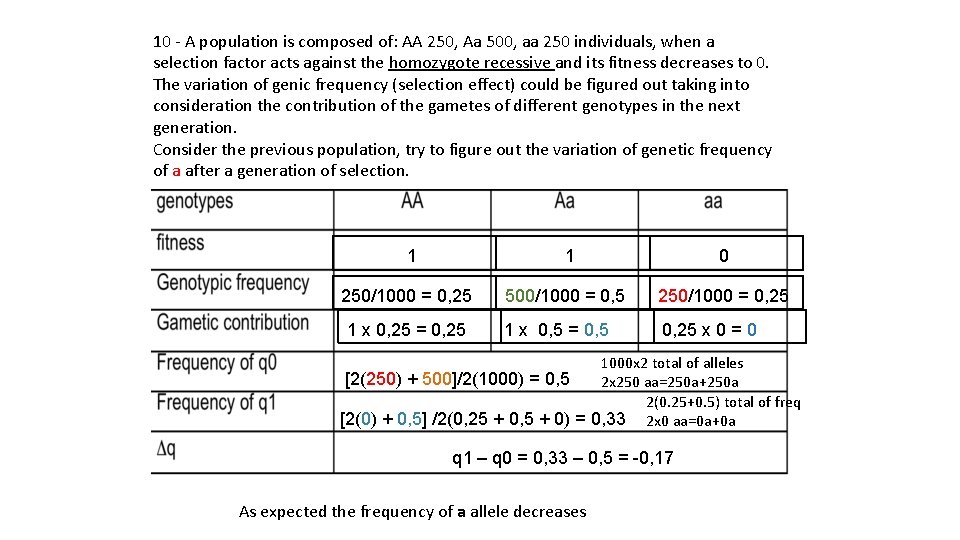
10 - A population is composed of: AA 250, Aa 500, aa 250 individuals, when a selection factor acts against the homozygote recessive and its fitness decreases to 0. The variation of genic frequency (selection effect) could be figured out taking into consideration the contribution of the gametes of different genotypes in the next generation. Consider the previous population, try to figure out the variation of genetic frequency of a after a generation of selection. 1 1 0 250/1000 = 0, 25 500/1000 = 0, 5 250/1000 = 0, 25 1 x 0, 25 = 0, 25 1 x 0, 5 = 0, 5 0, 25 x 0 = 0 1000 x 2 total of alleles [2(250) + 500]/2(1000) = 0, 5 2 x 250 aa=250 a+250 a 2(0. 25+0. 5) total of freq [2(0) + 0, 5] /2(0, 25 + 0, 5 + 0) = 0, 33 2 x 0 aa=0 a+0 a q 1 – q 0 = 0, 33 – 0, 5 = -0, 17 As expected the frequency of a allele decreases

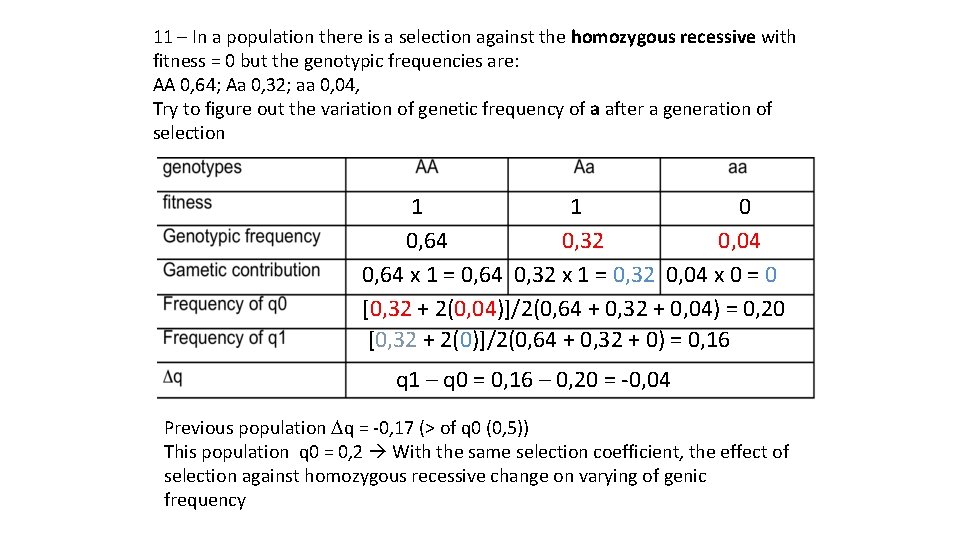
11 – In a population there is a selection against the homozygous recessive with fitness = 0 but the genotypic frequencies are: AA 0, 64; Aa 0, 32; aa 0, 04, Try to figure out the variation of genetic frequency of a after a generation of selection 1 0 0, 64 0, 32 0, 04 0, 64 x 1 = 0, 64 0, 32 x 1 = 0, 32 0, 04 x 0 = 0 [0, 32 + 2(0, 04)]/2(0, 64 + 0, 32 + 0, 04) = 0, 20 [0, 32 + 2(0)]/2(0, 64 + 0, 32 + 0) = 0, 16 q 1 – q 0 = 0, 16 – 0, 20 = -0, 04 Previous population Dq = -0, 17 (> of q 0 (0, 5)) This population q 0 = 0, 2 With the same selection coefficient, the effect of selection against homozygous recessive change on varying of genic frequency

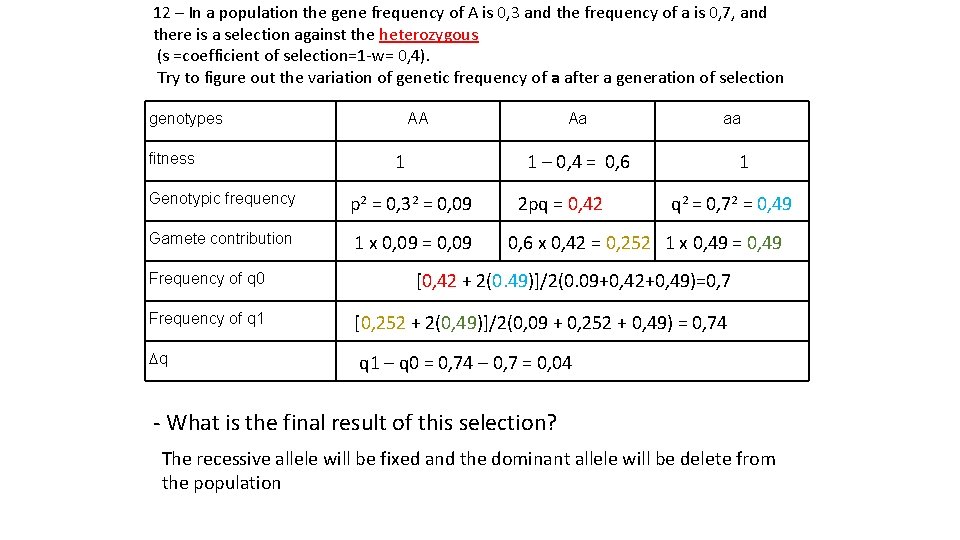
12 – In a population the gene frequency of A is 0, 3 and the frequency of a is 0, 7, and there is a selection against the heterozygous (s =coefficient of selection=1 -w= 0, 4). Try to figure out the variation of genetic frequency of a after a generation of selection genotypes AA Aa aa 1 – 0, 4 = 0, 6 1 fitness Genotypic frequency p 2 = 0, 32 = 0, 09 2 pq = 0, 42 q 2 = 0, 72 = 0, 49 Gamete contribution 1 x 0, 09 = 0, 09 0, 6 x 0, 42 = 0, 252 1 x 0, 49 = 0, 49 Frequency of q 0 [0, 42 + 2(0. 49)]/2(0. 09+0, 42+0, 49)=0, 7 Frequency of q 1 [0, 252 + 2(0, 49)]/2(0, 09 + 0, 252 + 0, 49) = 0, 74 Dq q 1 – q 0 = 0, 74 – 0, 7 = 0, 04 - What is the final result of this selection? The recessive allele will be fixed and the dominant allele will be delete from the population

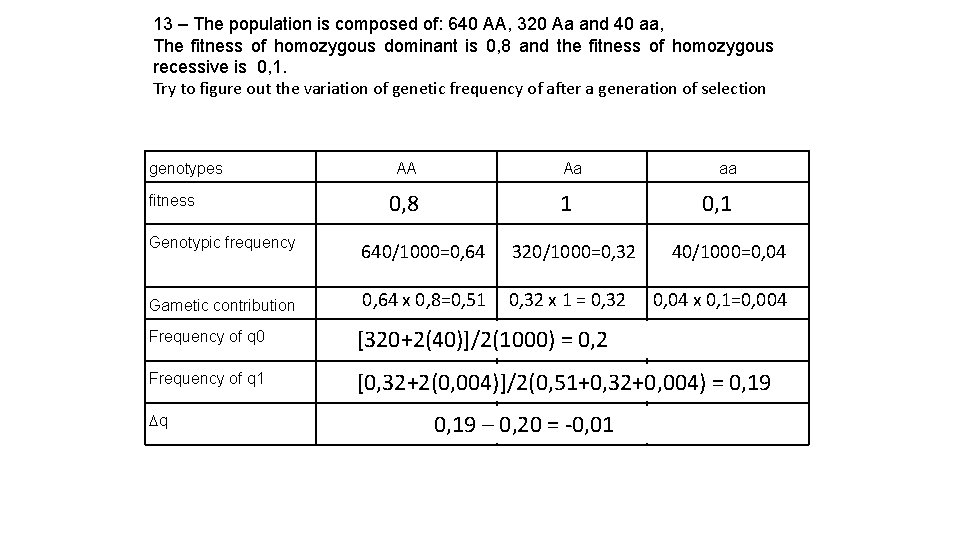
13 – The population is composed of: 640 AA, 320 Aa and 40 aa, The fitness of homozygous dominant is 0, 8 and the fitness of homozygous recessive is 0, 1. Try to figure out the variation of genetic frequency of after a generation of selection genotypes AA Aa aa 0, 8 1 0, 1 fitness Genotypic frequency Gametic contribution Frequency of q 0 [320+2(40)]/2(1000) = 0, 2 Frequency of q 1 [0, 32+2(0, 004)]/2(0, 51+0, 32+0, 004) = 0, 19 Dq 640/1000=0, 64 320/1000=0, 32 40/1000=0, 04 0, 64 x 0, 8=0, 51 0, 32 x 1 = 0, 32 0, 04 x 0, 1=0, 004 0, 19 – 0, 20 = -0, 01