Gel filtration Chromatography 1 Desalting group separation separate








- Slides: 24

Gel filtration Chromatography 1. Desalting (group separation): separate 2. the target protein from low-molecular mass contaminants. 2. Change buffer 3. protein fractionation

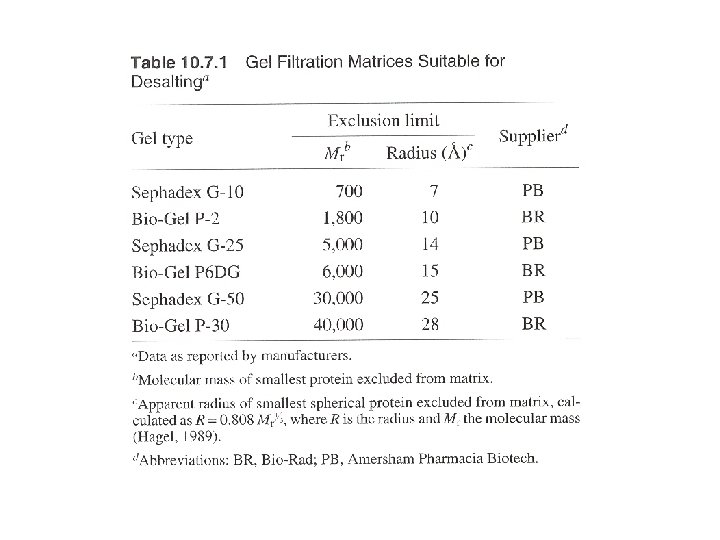
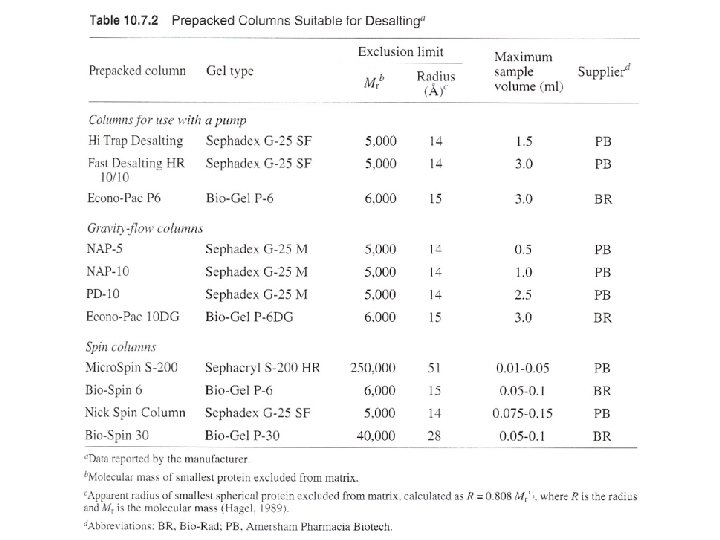
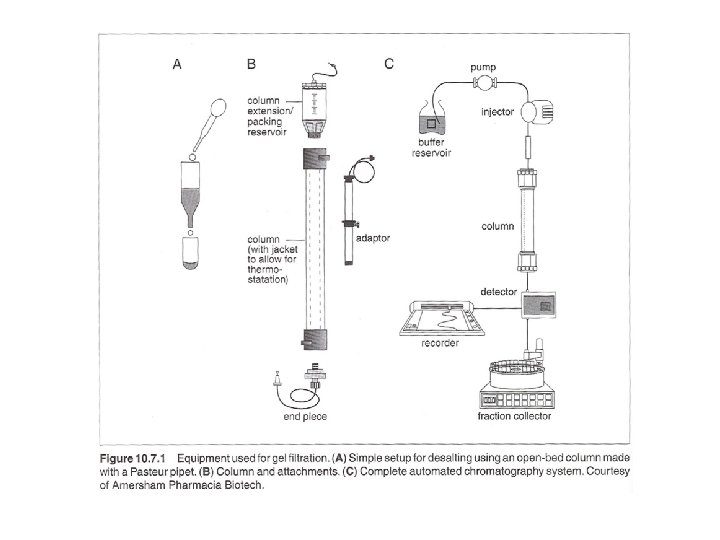
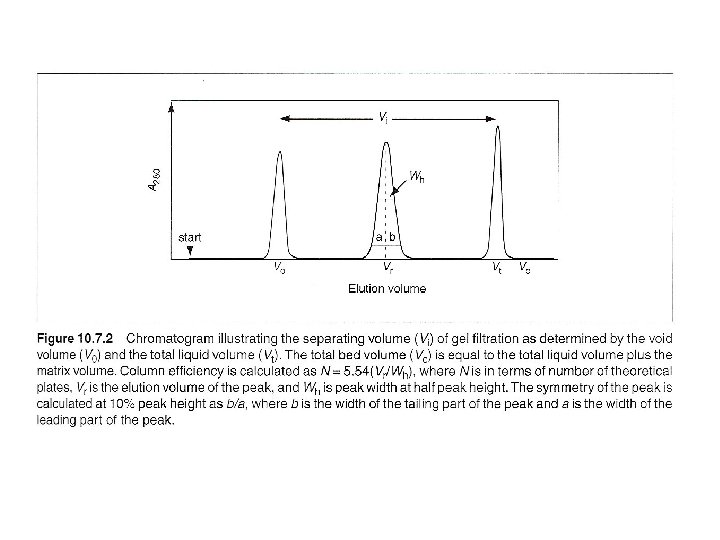
Three ways: columns with pump, gravity-flow columns, spin columns - Many pre-packed columns are available. - Can buy gel matrics, and prepare the gel, and pack the columns (10 -53 to 10 -57, or according to the suppliers’ suggestions). - Use Blue dextran 2000 (blue) or vitamine B 12 (yellow) as void volumn (V 0) marker. - Use acetone (5 mg/ ml) as total volumn (Vt) marker. (by absorbance at OD 280) - Gel matrx volumn + Vt = bed-volumn (Vc) - Vi (separating volumn) = Vt-V 0





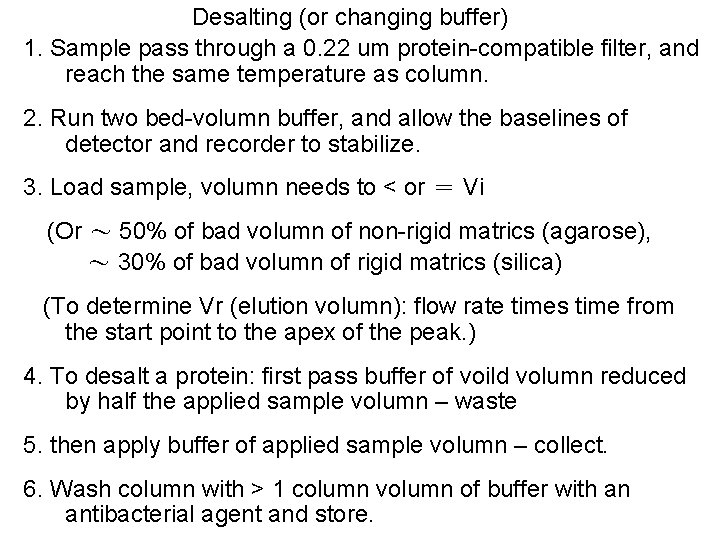
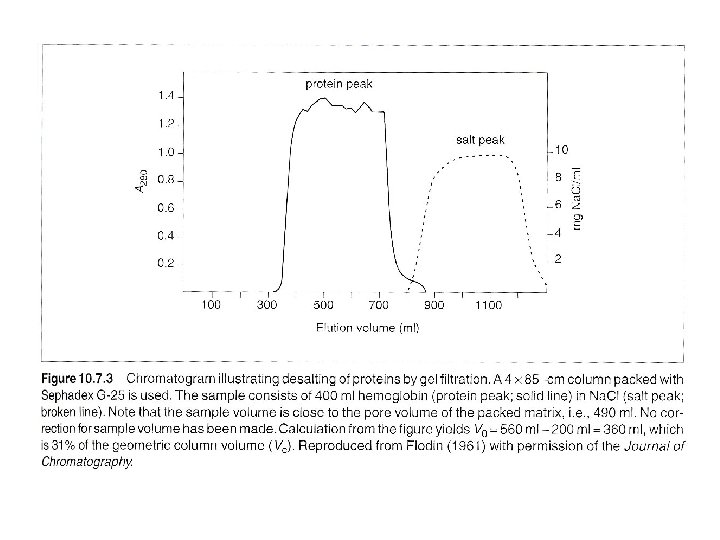
Desalting (or changing buffer) 1. Sample pass through a 0. 22 um protein-compatible filter, and reach the same temperature as column. 2. Run two bed-volumn buffer, and allow the baselines of detector and recorder to stabilize. 3. Load sample, volumn needs to < or = Vi (Or ~ 50% of bad volumn of non-rigid matrics (agarose), ~ 30% of bad volumn of rigid matrics (silica) (To determine Vr (elution volumn): flow rate times time from the start point to the apex of the peak. ) 4. To desalt a protein: first pass buffer of voild volumn reduced by half the applied sample volumn – waste 5. then apply buffer of applied sample volumn – collect. 6. Wash column with > 1 column volumn of buffer with an antibacterial agent and store.


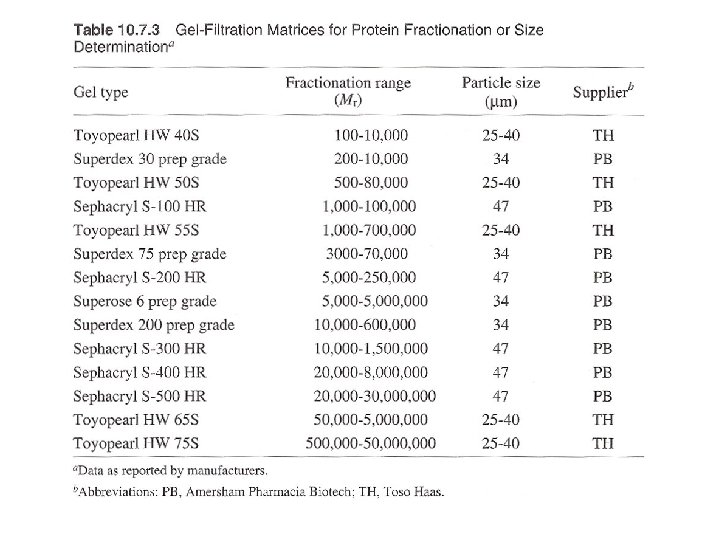
Gel filtration for protein fractionation (usually used in the polishing step in protein purification) - Columns are different from desalting - Sample volumn about 2% of the bed volumn. - Purity of the peak can be checked by HPLC or electrophotresis. - Proteins molecular weight could be determined by Vr (elution volumn). But mass spectrophotometry (MS) can do better job in determination of molecular mass.





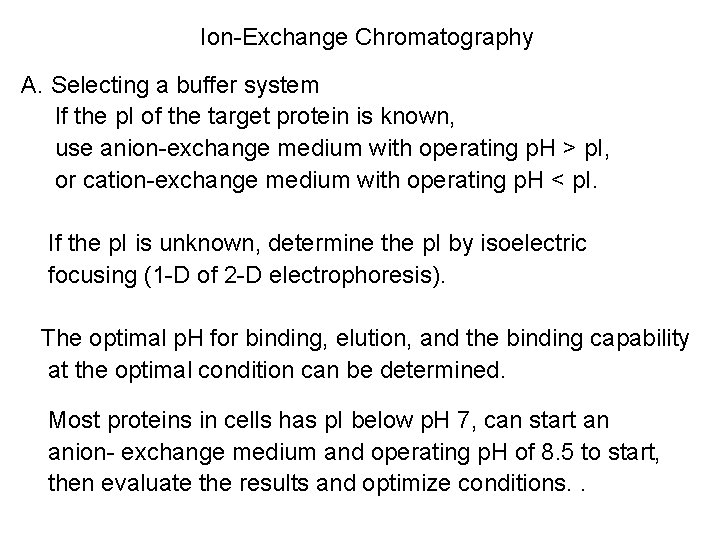
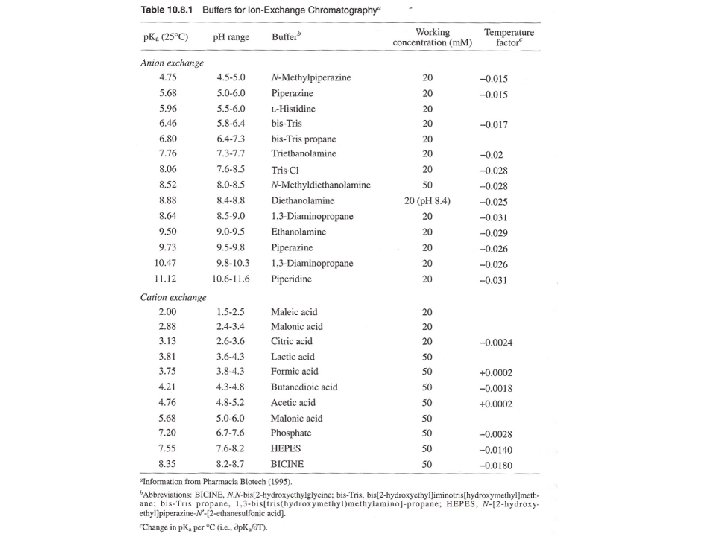
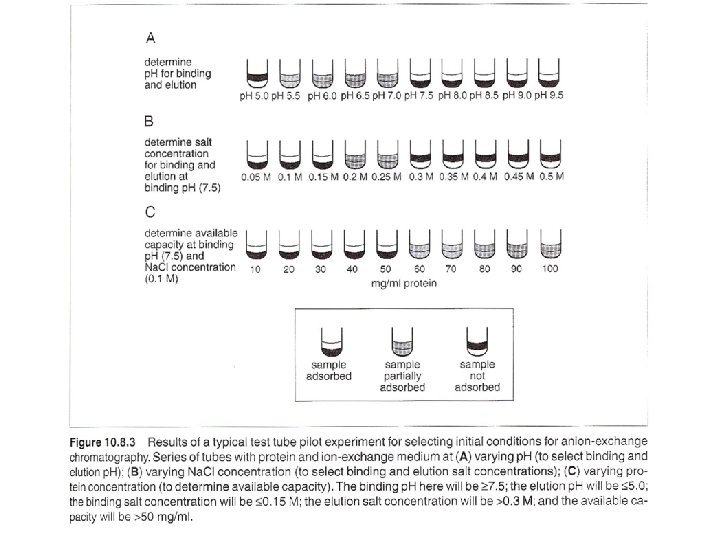
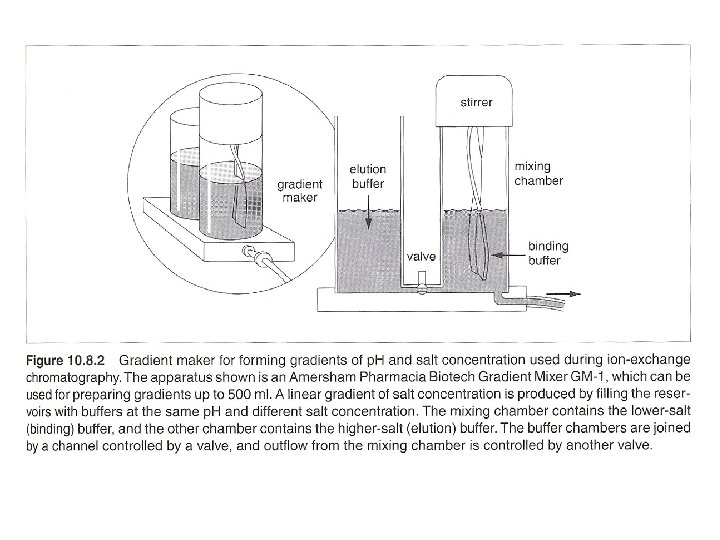
Ion-Exchange Chromatography A. Selecting a buffer system If the p. I of the target protein is known, use anion-exchange medium with operating p. H > p. I, or cation-exchange medium with operating p. H < p. I. If the p. I is unknown, determine the p. I by isoelectric focusing (1 -D of 2 -D electrophoresis). The optimal p. H for binding, elution, and the binding capability at the optimal condition can be determined. Most proteins in cells has p. I below p. H 7, can start an anion- exchange medium and operating p. H of 8. 5 to start, then evaluate the results and optimize conditions. .

B. Selecting a buffer system - Consider the p. H stability of the sample. - Anionic buffers for cation exchange, - Cationic buffers for anion exchange. (Thus, buffering ions will not bind gel matrix)



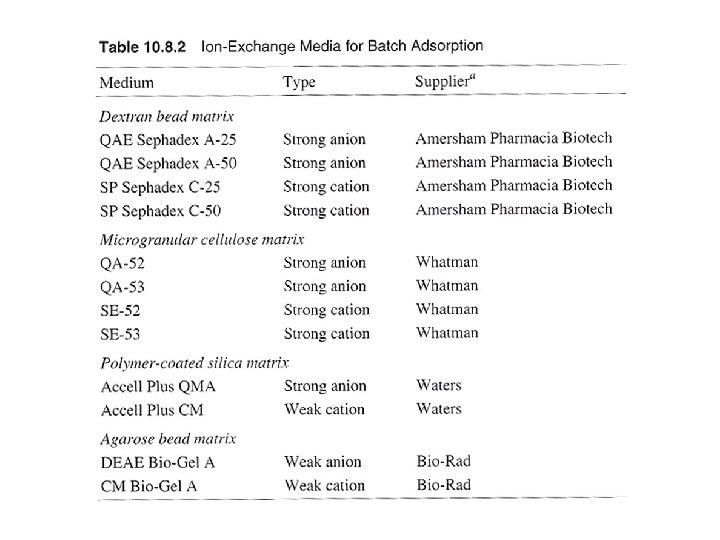
Batch adsorption and step-gradient elution (with increasing salt concentration) eg. anion exchange gel, binding buffer: 20 m. M Tris. Cl, p. H 7. 5 washing buffer: 20 m. M Tris. Cl, p. H 7. 5/100 m. M Na. Cl elution buffer: 20 m. M Tris. Cl, p. H 7. 5/350 m. M Na. Cl regeneration buffer: 20 m. M Tris. Cl, p. H 7. 5/ 2 M Na. Cl


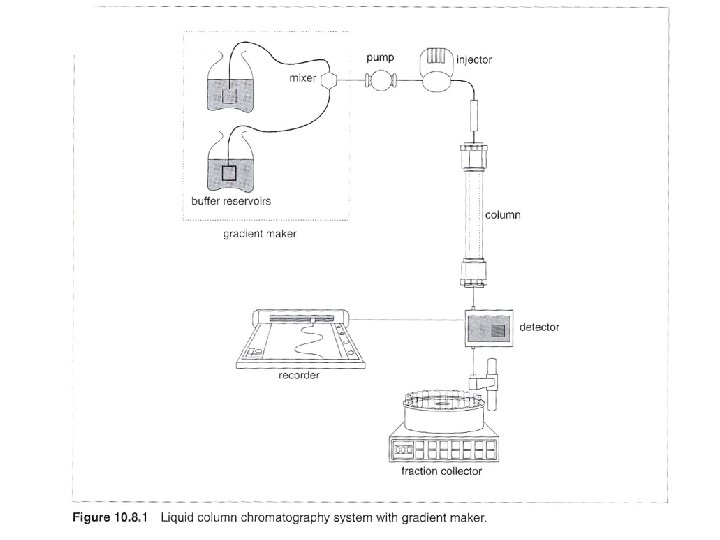
Column chromatography with linear gradient elution eg. 1 ml bed-volumn anion exchange column, binding buffer: 20 m. M Tris. Cl, p. H 7. 5 elution buffer: 20 m. M Tris. Cl, p. H 7. 5/ 1 M Na. Cl 1. Wash column: run 5 Vc volumn of elution buffer at 5 ml/min. 2. Equilibrium: run 5 -10 Vc volumn of binding buffer at 5 ml/min. 3. Collect one fraction at the end, measure p. H and conductivity, see if the same as binding buffer. 4. Filter sample, use total proteins of 25 mg for initial loading. 0. 22 u. M filter for beads < 34 um, 0. 45 um filter for beads between 34 um and 90 um, 1 um filter for beads > 90 um

5. Open injection valve for sample injection, and begin to collect fractions at 1 ml. Can reduce flow rate for very concentrated sample, or increase flow rate for very diluted sample. 6. After sample injected, wash with 3 to 5 Vc volumn of binding buffer at flow rate 5 ml/min. Monitor signal to baseline. 7. Close sample injection valves to reduce system dead volumn. 8. Elute with a linear gradient from 0% to 100% elution buffer in 20 Vc (20 ml). 9. Regenerate colunmn by washing with 5 Vc of elution buffer. 10. Reequilibrate column with 5 to 10 Vc of binding buffer.



Preparation of antibody-sepharose Covalently linking an antibody to Sepharose (CL-4 B, or CL-2 b for high MW antigen), using CNBr activation method. 1. dialyze 1 -30 m g/ml antibody against 0. 1 M Na. HCO 3 /0. 5 M Na. Cl at 40 C with 3 buffer changes over 24 hrs, use dialysis solutions 500 times of the antibody volumn. 2. Centrifuge 3. Measure A 280 (mg/ml Ig. G =A 280 / 1. 44), dilute to 5 mg/ml with 0. 1 M Na. HCO 3 /0. 5 M Na. Cl 4. Wash the sepharose with 10 vol water, use Waterman no. 1 filter and Buchner funnel. 5. Add equal volumn of 0. 2 M Na 2 CO 3 to Sepharose. 6. Add CNBr/ acetonitrile dropwise (in hood). 7. Filter, dry, and add 0. 1 M HCl, and add antibody solution, for 2 hr at rt. 8. Add glycine to saturate the active group on Sepharose, and measure A 280 to determine percentage coupling
 Gel filtration chromatography lab
Gel filtration chromatography lab Advantages of gel filtration chromatography
Advantages of gel filtration chromatography Gel filtration chromatography definition
Gel filtration chromatography definition The inlet pressure in a constant rate filtration
The inlet pressure in a constant rate filtration Filtration is used to separate
Filtration is used to separate What is void volume in gel filtration
What is void volume in gel filtration Kav gel filtration
Kav gel filtration Gel filtration application
Gel filtration application Principle of size exclusion chromatography
Principle of size exclusion chromatography Purification table
Purification table Agarose gel vs polyacrylamide gel
Agarose gel vs polyacrylamide gel Agarose gel vs polyacrylamide gel
Agarose gel vs polyacrylamide gel Gel kim olursan ol yine gel
Gel kim olursan ol yine gel Chromatography is a technique used to separate
Chromatography is a technique used to separate Gel chromatography instrumentation
Gel chromatography instrumentation Filtration separation solution
Filtration separation solution Paper chromatography separation of cations and dyes
Paper chromatography separation of cations and dyes Pixabay
Pixabay Peritubular capillaries
Peritubular capillaries Autoregulation of gfr
Autoregulation of gfr What is the process called
What is the process called Conclusion sur la distillation
Conclusion sur la distillation Distillation and decantation
Distillation and decantation Can sulfur and water be separated by filtration
Can sulfur and water be separated by filtration Filtration fraction
Filtration fraction