Eukaryotic Comparative Genomics June 2018 GEP Alumni Workshop






































- Slides: 40

Eukaryotic Comparative Genomics June 2018 GEP Alumni Workshop Last Update: 12/23/2020 Barak Cohen

Detecting Conserved Sequences Charles Darwin Motoo Kimura

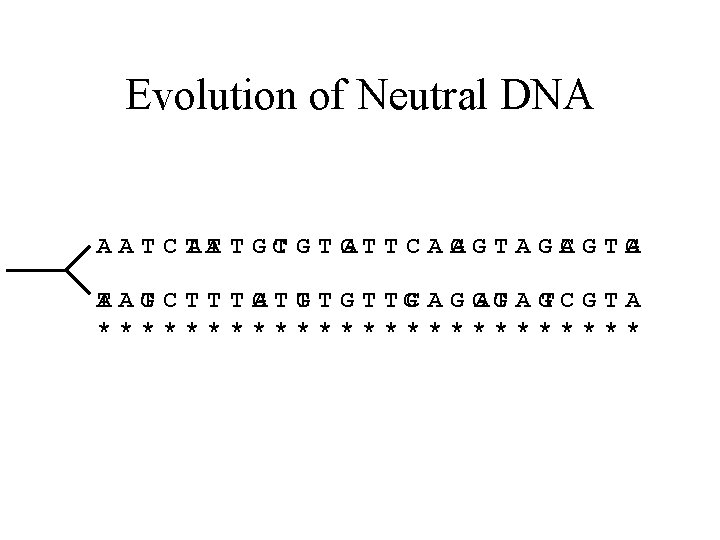
Evolution of Neutral DNA AATCT AA TTGC T GA T T C A G AGTAGC AGTG A TAG A TCTTTG ATG T T GC A G GA G T A GT C G T A *************

Evolution of Non-Neutral DNA AT CTA GT C C GA T GC GTACCGACCATAA GGAT GC AC A CG TATA CCATGTGGTAT CCGA TC C A T A A GC ATAT C ***************

Multi-Species Alignment ATGTGGCGCAGCCTGTGCCAGCTGGACGATCGA ATGTAGCCAGTGCCAGCTGGACGATCGA GTACATCGATAGCTTAGAATGCTGGACGATCTC GTACGTCGATAGCATAGAATGCTGGACGATCTC * * ******

How to do Comparative Genomics 1. Choose species to analyze 2. Align sequences 3. Identify streches of highly conserved nucleotides

Choose species closely related species • Closely Related Species – align well – not many changes distantly related species • Distantly Related Species – hard to align – lots of changes

~10 Mya ~20 Mya S. cerevisiae S. cariocanus S. paradoxus S. mikatae S. kudriavzevii S. bayanus S. pastorianus S. servazzii S. unisporus S. exiguus S. diarenensis S. castellii S. kluyveri ~150 Mya >350 Mya Kluyveromyces lactis Schizosaccharomyces pombe

Case Study: Coding vs. Non-Coding ATG…. • Non-Coding DNA - regulatory functions - short (5 -15 bp) - degenerate - variable spacing ORF …TAA • Coding DNA - codes for protein - triplet code - open reading frame (ORF) - tend to be long (50 -500 bp) - highly constrained

CASE 1: Non-Coding ATG… GAL 4 …TAA

~10 Mya ~20 Mya S. cerevisiae S. cariocanus S. paradoxus S. mikatae S. kudriavzevii S. bayanus S. pastorianus S. servazzii S. unisporus S. exiguus S. diarenensis S. castellii S. kluyveri ~150 Mya >350 Mya Kluyveromyces lactis Schizosaccharomyces pombe

Closely-related sequences are uninformative ATG… GAL 4 paradoxus cerevisiae TCTTCTGAGACAGCATCACTTCTTCTTNTTTTTTACATAACTTATTCTTCTATAATTTTC TCCTTTGAGACAGCATTCGCCCAGTATTTTATTCTACA-AACCTTCTATAATTT-C ** * ******* ****** * paradoxus cerevisiae AACGTATTTACATAGTTCTGTATCAGTTTAATCACCATAATATTGTTTTCCCTCAACTAA AAAGTATTTACATAATTCTGTATCAGTTTAATCACCATAATATCGTTTTCT-----TTGT ** ******************* * paradoxus cerevisiae TGAATGCAATTAGATTTTCTTATTGTTCCCTCGCGGCTTTTGTTTTATAATCTATT TTAGTGCAATTTTTCCTATTGTTACTTCG-GGCCTTTTTCTGTTTTATGAGCTATT * * ******* ******** * ***** paradoxus cerevisiae TTTTCCGTCATTTCTTCCCCAGATTTCCAACTTCATCTCCAGATTGTGTCTATGTAATGC TTTTCCGTCATC-CTTCCCCAGATTTTCAGCTTCATCTCCAGATTGTGTCTACGTAATGC ************* ******* paradoxus cerevisiae ATGCTATCATATTGAGAAAAGATAGAGAAACAACCCTCCTGAAAAATGAAGCTACTGTCT ACGCCATCATTTTAAGAGAGGACAGAGAAGCCTCCTGAAAGATGAAGCTACTGTCT * ** ***** ** ****** ********

~10 Mya ~20 Mya S. cerevisiae S. cariocanus S. paradoxus S. mikatae S. kudriavzevii S. bayanus S. pastorianus S. servazzii S. unisporus S. exiguus S. diarenensis S. castellii S. kluyveri ~150 Mya >350 Mya Kluyveromyces lactis Schizosaccharomyces pombe

Distantly-related sequences do not align ATG… GAL 4 Noncoding (Promoter) cerevisiae ACTTACCAT-CAAC-CATAGATGGGTAAAC---GGTTAGTAACTAGGAACACGAT castelli AGA-GTCAAACTTTTCGT—ATA--TATAATATGTCTGATTGCTGGTT---T * ** * * * *

~10 Mya ~20 Mya S. cerevisiae S. cariocanus S. paradoxus S. mikatae S. kudriavzevii S. bayanus S. pastorianus S. servazzii S. unisporus S. exiguus S. diarenensis S. castellii S. kluyveri ~150 Mya >350 Mya Kluyveromyces lactis Schizosaccharomyces pombe

Multiple sequence alignments reveal conserved elements ATG… cerevisiae mikatae Bayanus kudriadzevi GAL 4 TGAGACAGCAT-CACTTCTT-CTTNTTTTTTACATAACTTATTCTTCTATAATTTTCAAC TGAGACAGCATTCACTTCTTTTTACATATCTTATTCTTCTATAATTTTCAAC TGAGACAGCATTCGCCCAGT--ATTTTAT-TCTACAAACCTTCTATAATTT-CAAA TGAGACTGCACTCCC----TCTTCCTTTC------TCCATAACTT---AC ****** * * * ** **** ** * UAS 1 UAS 2 paradoxus kluyveri cerevisiae bayanus GTATTTACATAGTTCTGTATCAGTTTAATCACCATAAT------ATTGTTTTCCCTCAAC GTATTTACATAATTCTGTATCAGTTTAATCACCATAAT------ATCGTTTTCTTTGT-TTATTTACATAGTTTTGTATCAGTTTAATCACCATAATCGTAACACCGTTTTACCTCACC ***** ** ************ * paradoxus kluyveri cerevisiae bayanus TAATGCAATTAGATTTTC-TTATTGTTCCC-TCGCGGCTTTTGTTTTATAATGCAATTAGATTTTCCTTATTGTTCCCCTCGCGGCTTTTGTTTTATAAT ---TTAGTGCAATTTTTC-CTATTGTTACT-TCG-GGCCTTTTTCTGTTTTATGAG TGATGCGGG--A---ATCCTTC-AGACCGTTCTC-TCGCGC---------* * * *** *** * UES MIG 1 paradoxus kluyveri cerevisiae bayanus -CTATTTTTTCCGTCATTTCTTCCCC-AGATTTCCAACTTCAT-CTCCAGATTGTGTCTA ACTATTTTTTCCGTCATTTCTTCCCCCAGATTTCCAACTTCATACTCCAGATTGTGTCTA -CTATTTTTTCCGTCATC-CTTCCCC-AGATTTTCAGCTTCAT-CTCCAGATTGTGTCTA -CTTTTTCGTCATTTCTTCCCC-AGATCTACAACTTTAA-CTCCAGACGGTGTATA ** ******* * ******* ** paradoxus kluyveri cerevisiae bayanus TGTAATGCATGCTATCATATTGAGAAAAGATAGAGAAACAACCCTCCTGAAAAATGAAGC CGTAATGCACGCCATCATTTTAAGAGAGGACAGAGAAGCCTCCTGAAAGATGAAGC GGCAGTACAAGCAGTGCTTTTGGGAAGAGGCAAAGCTGCAGACCTCGAGAACAATGAAGC * ** ** *** *******

CASE 2: Coding ATG… CLN 3 …TAA

~10 Mya ~20 Mya S. cerevisiae S. cariocanus S. paradoxus S. mikatae S. kudriavzevii S. bayanus S. pastorianus S. servazzii S. unisporus S. exiguus S. diarenensis S. castellii S. kluyveri ~150 Mya >350 Mya Kluyveromyces lactis Schizosaccharomyces pombe

Closely-related sequences are uninformative

~10 Mya ~20 Mya S. cerevisiae S. cariocanus S. paradoxus S. mikatae S. kudriavzevii S. bayanus S. pastorianus S. servazzii S. unisporus S. exiguus S. diarenensis S. castellii S. kluyveri ~150 Mya >350 Mya Kluyveromyces lactis Schizosaccharomyces pombe

Less distantly related species not informative either

~10 Mya ~20 Mya S. cerevisiae S. cariocanus S. paradoxus S. mikatae S. kudriavzevii S. bayanus S. pastorianus S. servazzii S. unisporus S. exiguus S. diarenensis S. castellii S. kluyveri ~150 Mya >350 Mya Kluyveromyces lactis Schizosaccharomyces pombe

Distantly-related species reveal functional protein domains

Identification of Multi-Species Conserved Regions (MCS) Human Chimp Mouse Rat Dog cccattcttttccaagtgtctccg--cctgcagcgattaggttagaaagcatttctctct ttcagtcgtttcccagtgtctctga-cattcagagactactttagtaagcattt-tctct tcagtccttccctggcatctccag-cactcaa-gactactttagtaagcattt-tctctg tcaatgactttcccagtctcttctactgggaagagattaggttgcaaatcatttttctct * * * ** How can we decide if this region is “conserved? ” Margulies et al (2003) Gen. Res. 13: 2507 -18

Its like flipping coins (really)

Binomial-Based Method for Detecting Conserved Sequences Human: AATGG Mouse: AATCG Status: CCCDC p = probability that a site is the same between human and mouse by chance alone (Kimura), q = 1 -p For an alignment N base pairs long with n identities calculate the cumulative binomial probability as: Margulies et al (2003) Gen. Res. 13: 2507 -18



Large sequencing projects are underway

Tree Topology Influences Power Star Phylogeny species A species F species B species E species D species C Actual Phylogeny

Challenges in larger genomes 1) Deciding on the neutral rate of substitution 2) Local differences in neutral rate of substitutions 3) Multiple hypothesis testing 4) Repeat sequences and uneven base composition

Phast. Cons and the UCSC Genome Browser OLIG 2 100 kb upstream of OLIG 2

Motif Searching Across Several Multiple Alignments Gene 1 Species 2 Species 3 Gene 2 Gene N Gene 3 …

Information Content Eco. R 1 Random Rap 1 GAATTC GAATTC GCCTAC ACATTC TCATTC CGACTC GAATTC ATATCG GAAATG TGTATGGGTG TGTTCGGATT TGCATGGGTG TGTACAGGTG TGTATGGATG TGTTCGGGTT TGTATGGGTG

Weight Matrix Model of TATA Box A: -8 10 -1 2 1 -8 C: -10 -9 -3 -2 -1 -12 G: -7 -9 -1 -1 -4 -9 T: 10 -6 9 0 -1 11 G. Stormo

Weight Matrix Model of TATA Box Score = -24 C T A: -8 10 -1 C: -10 -9 G: -7 T: 10 …. A A T A A 2 1 -8 -3 -2 -1 -12 -9 -1 -1 -4 -9 -6 9 0 -1 11 T G T… G. Stormo

Weight Matrix Model of TATA Box Score = 43 …. A T A A: -8 10 C: -10 G: T: C T A A T -1 2 1 -8 -9 -3 -2 -1 -12 -7 -9 -1 -1 -4 -9 10 -6 9 0 -1 11 G T… G. Stormo

Weight Matrix Model of TATA Box N(b, i) F(b, i) S(b, i) = log[F(b, i)/P(b)] G. Stormo

Now we can compare motifs to each other A C G T 4 -3 5 -6 -2 -5 2 -1 -2 11 -1 -1 -10 8 2 -4 2 -3 -3 2 1 2 -3 15 A C G T 3 -2 2 1 3 -1 -2 7 -2 -1 -8 6 3 -2 2 -2 -1 1 1 4 -3 9

MAGMA unaligned motif finding in multispecies conserved regions Gene 1 Species 2 Species 3 Gene 2 Gene N Gene 3 … *Ihuegbu, Stormo, & Buhler, JCB 19: 139, 2012