ENZYMES A protein with catalytic properties due to






- Slides: 33

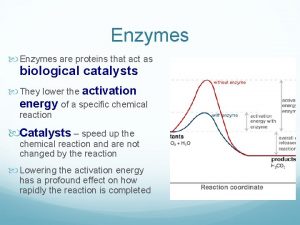
ENZYMES A protein with catalytic properties due to its power of specific activation © 2007 Paul Billiet ODWS

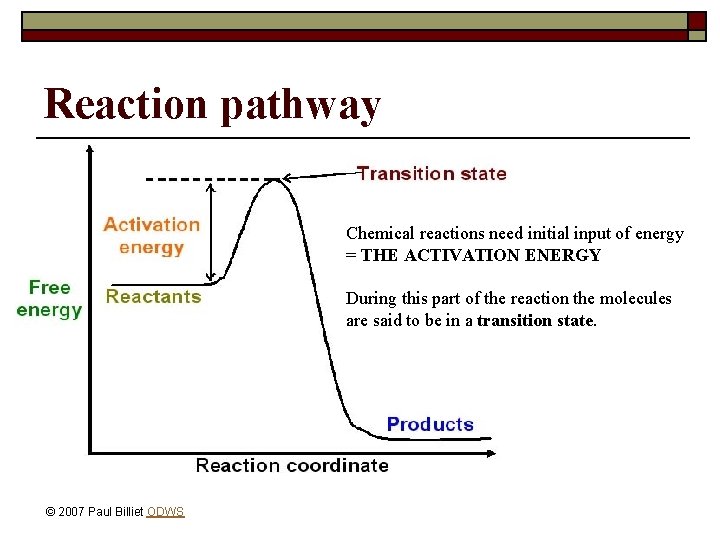
Reaction pathway Chemical reactions need initial input of energy = THE ACTIVATION ENERGY During this part of the reaction the molecules are said to be in a transition state. © 2007 Paul Billiet ODWS

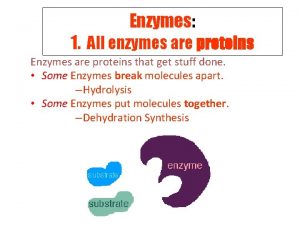
Enzymes Modify Chemical Reactions o o o Reactions with enzymes are faster Reactions with enzymes are more specific Enzymes increase rate of reactions without increasing temperature. They do this by lowering activation energy. They create a new reaction pathway “a short cut” © 2007 Paul Billiet ODWS (edited by ckelly 2014)

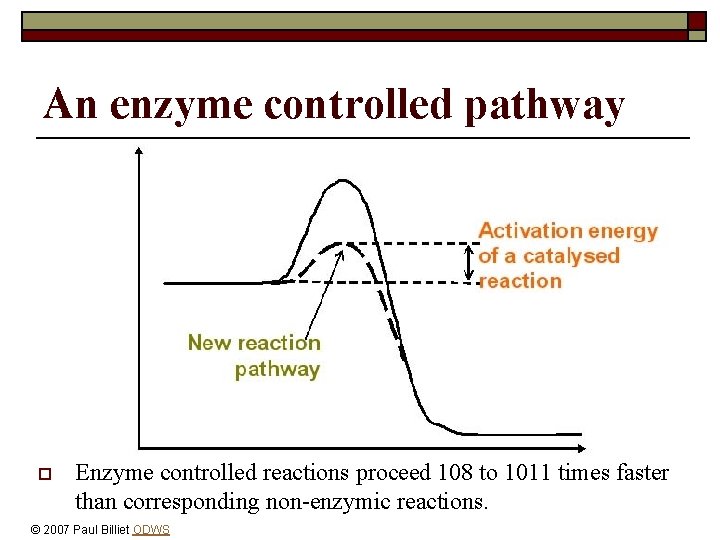
An enzyme controlled pathway o Enzyme controlled reactions proceed 108 to 1011 times faster than corresponding non-enzymic reactions. © 2007 Paul Billiet ODWS

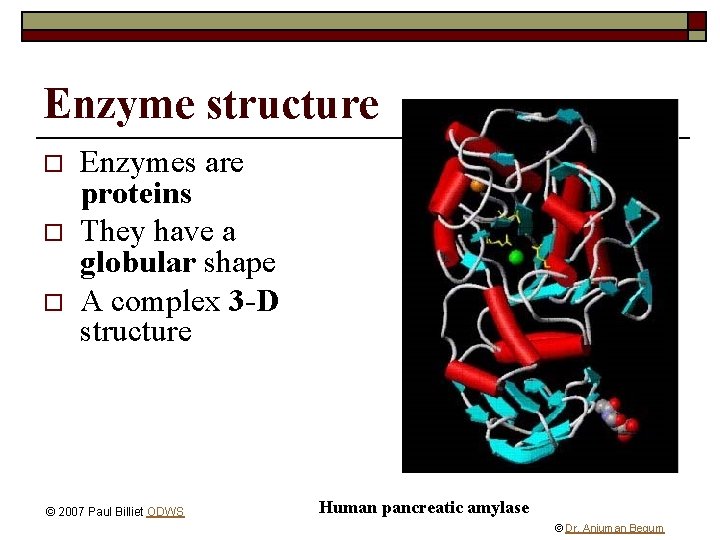
Enzyme structure o o o Enzymes are proteins They have a globular shape A complex 3 -D structure © 2007 Paul Billiet ODWS Human pancreatic amylase © Dr. Anjuman Begum

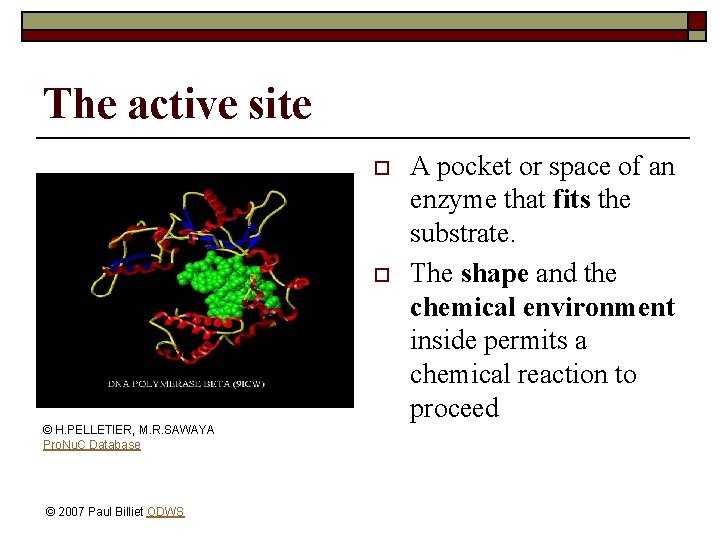
The active site o o © H. PELLETIER, M. R. SAWAYA Pro. Nu. C Database © 2007 Paul Billiet ODWS A pocket or space of an enzyme that fits the substrate. The shape and the chemical environment inside permits a chemical reaction to proceed

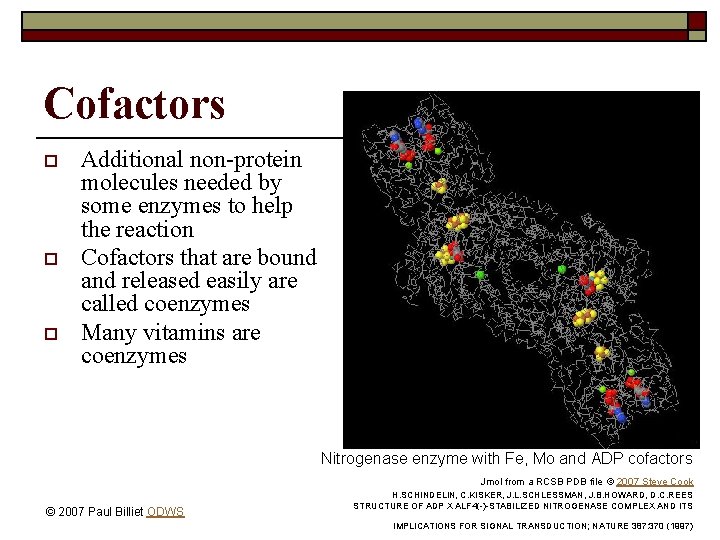
Cofactors o o o Additional non-protein molecules needed by some enzymes to help the reaction Cofactors that are bound and released easily are called coenzymes Many vitamins are coenzymes Nitrogenase enzyme with Fe, Mo and ADP cofactors Jmol from a RCSB PDB file © 2007 Steve Cook © 2007 Paul Billiet ODWS H. SCHINDELIN, C. KISKER, J. L. SCHLESSMAN, J. B. HOWARD, D. C. REES STRUCTURE OF ADP X ALF 4(-)-STABILIZED NITROGENASE COMPLEX AND ITS IMPLICATIONS FOR SIGNAL TRANSDUCTION; NATURE 387: 370 (1997)

The substrate o o o The reactants are changed by the enzyme Enzymes are specific to their substrates Specificity is determined by the active site © 2007 Paul Billiet ODWS

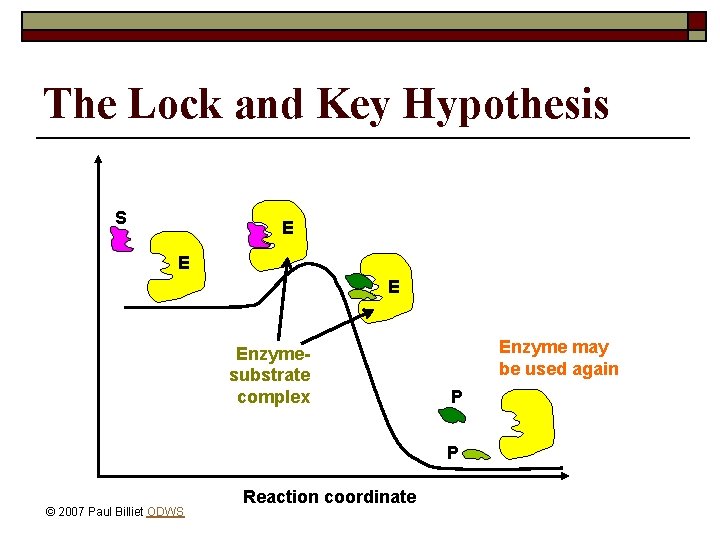
The Lock and Key Hypothesis o o o Fit between substrate and active site of enzyme is exact Temporary structure called enzyme-substrate complex forms Products have a different shape from the substrate Once formed, they are released from the active site Enzyme is free to act on another substrate © 2007 Paul Billiet ODWS

The Lock and Key Hypothesis S E Enzymesubstrate complex Enzyme may be used again P P © 2007 Paul Billiet ODWS Reaction coordinate

The Lock and Key Hypothesis o o This explains enzyme specificity This explains the loss of activity when enzymes denature © 2007 Paul Billiet ODWS

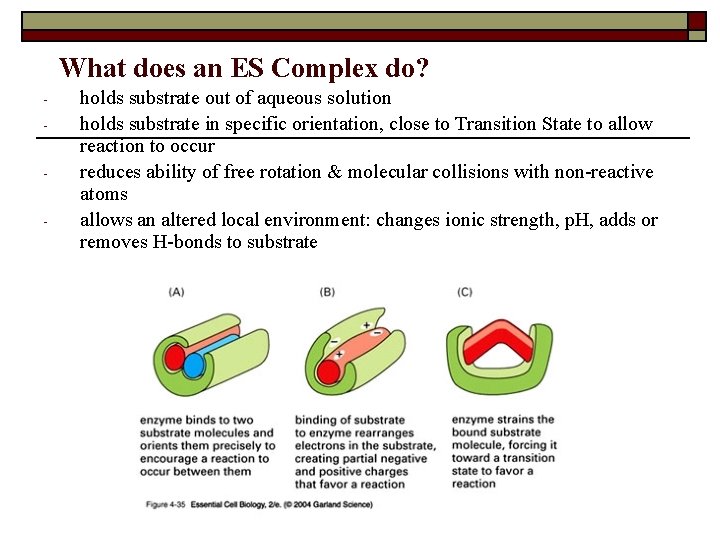
What does an ES Complex do? - holds substrate out of aqueous solution holds substrate in specific orientation, close to Transition State to allow reaction to occur reduces ability of free rotation & molecular collisions with non-reactive atoms allows an altered local environment: changes ionic strength, p. H, adds or removes H-bonds to substrate

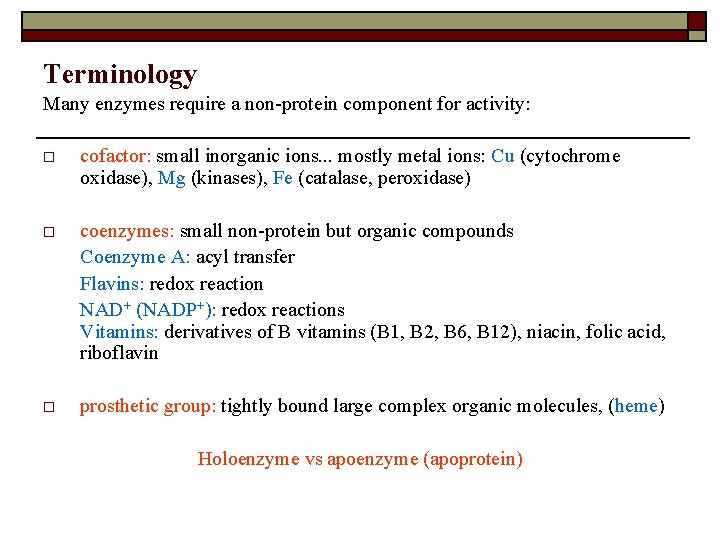
Terminology Many enzymes require a non-protein component for activity: o cofactor: small inorganic ions. . . mostly metal ions: Cu (cytochrome oxidase), Mg (kinases), Fe (catalase, peroxidase) o coenzymes: small non-protein but organic compounds Coenzyme A: acyl transfer Flavins: redox reaction NAD+ (NADP+): redox reactions Vitamins: derivatives of B vitamins (B 1, B 2, B 6, B 12), niacin, folic acid, riboflavin o prosthetic group: tightly bound large complex organic molecules, (heme) Holoenzyme vs apoenzyme (apoprotein)

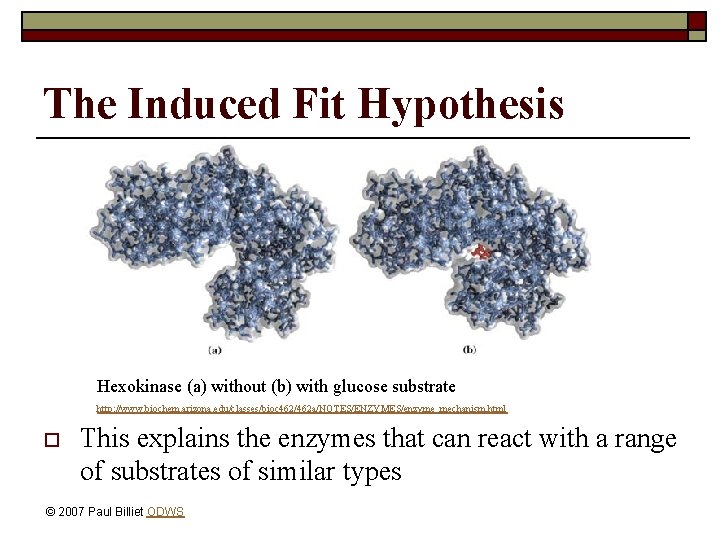
The Induced Fit Hypothesis o o Some proteins change their shape when binding substrate The bonds of the substrate are stretched to make the reaction easier (lowers activation energy) © 2007 Paul Billiet ODWS

The Induced Fit Hypothesis Hexokinase (a) without (b) with glucose substrate http: //www. biochem. arizona. edu/classes/bioc 462/462 a/NOTES/ENZYMES/enzyme_mechanism. html o This explains the enzymes that can react with a range of substrates of similar types © 2007 Paul Billiet ODWS

Factors affecting Enzymes o o substrate concentration p. H temperature inhibitors © 2007 Paul Billiet ODWS

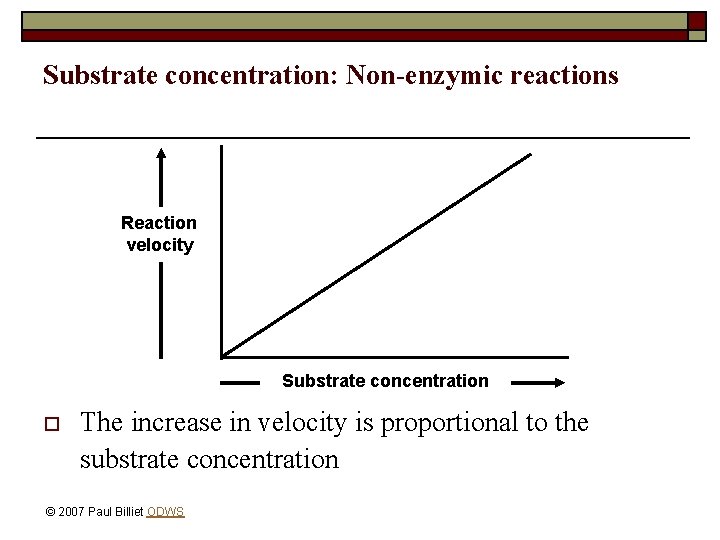
Substrate concentration: Non-enzymic reactions Reaction velocity Substrate concentration o The increase in velocity is proportional to the substrate concentration © 2007 Paul Billiet ODWS

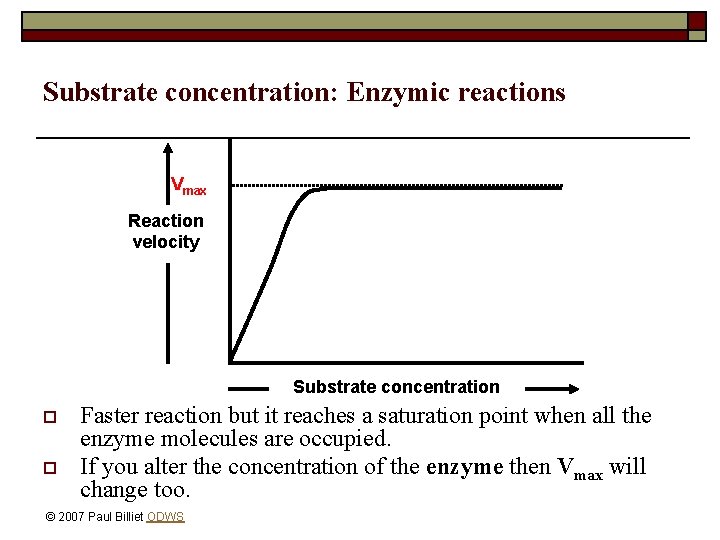
Substrate concentration: Enzymic reactions Vmax Reaction velocity Substrate concentration o o Faster reaction but it reaches a saturation point when all the enzyme molecules are occupied. If you alter the concentration of the enzyme then Vmax will change too. © 2007 Paul Billiet ODWS

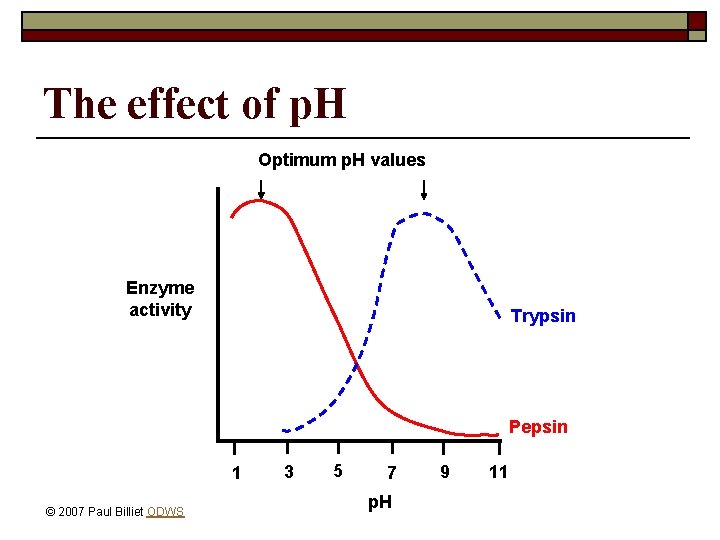
The effect of p. H Optimum p. H values Enzyme activity Trypsin Pepsin 1 © 2007 Paul Billiet ODWS 3 5 7 p. H 9 11

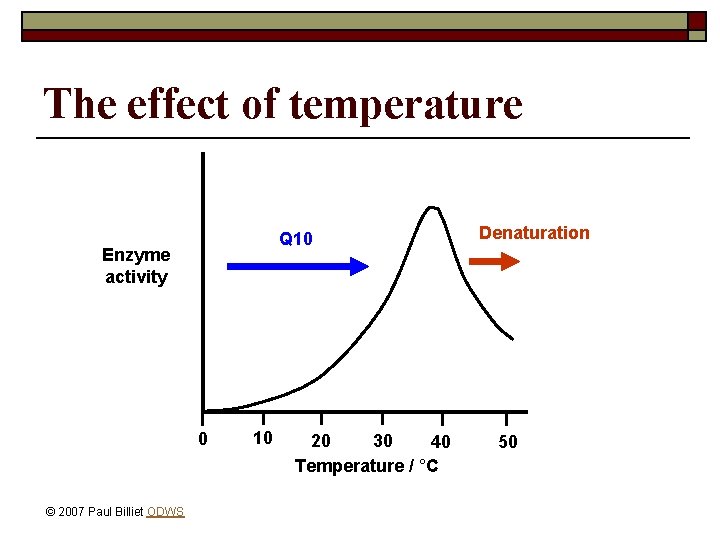
The effect of temperature Q 10 Enzyme activity 0 © 2007 Paul Billiet ODWS 10 20 30 40 Temperature / °C Denaturation 50

Inhibitors o o Reduce the rate of enzymatic reactions. Usually specific and work at low concentrations. Block the enzyme but they do not usually destroy it. Many drugs and poisons are inhibitors of enzymes in the nervous system. © 2007 Paul Billiet ODWS

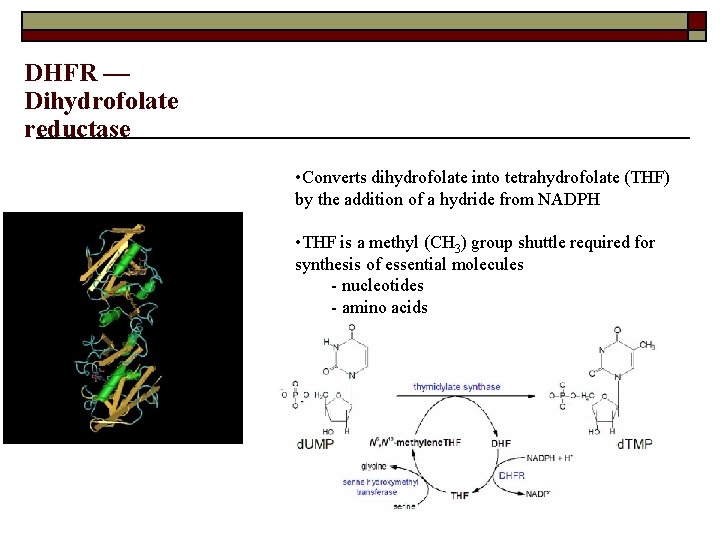
DHFR — Dihydrofolate reductase • Converts dihydrofolate into tetrahydrofolate (THF) by the addition of a hydride from NADPH • THF is a methyl (CH 3) group shuttle required for synthesis of essential molecules - nucleotides - amino acids 22

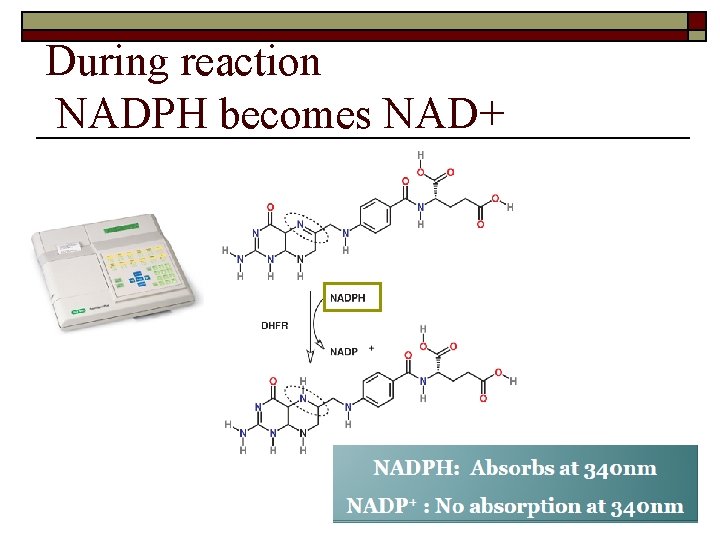
During reaction NADPH becomes NAD+ 23

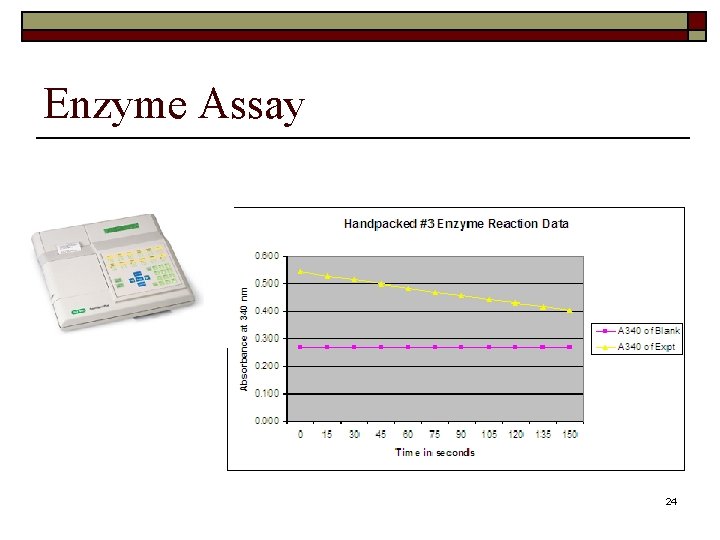
Enzyme Assay 24

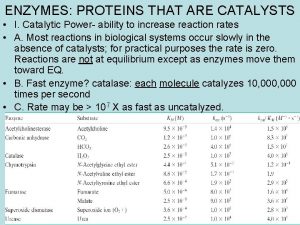
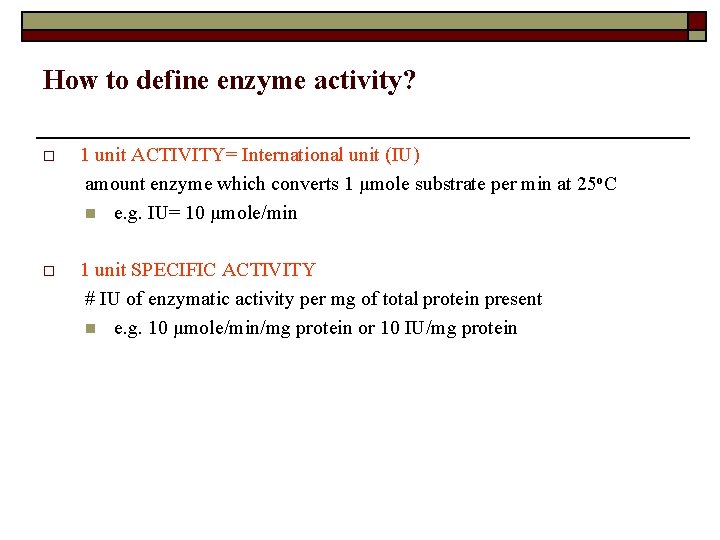
How to define enzyme activity? o 1 unit ACTIVITY= International unit (IU) amount enzyme which converts 1 μmole substrate per min at 25 o. C n e. g. IU= 10 μmole/min o 1 unit SPECIFIC ACTIVITY # IU of enzymatic activity per mg of total protein present n e. g. 10 μmole/min/mg protein or 10 IU/mg protein

Tools of enzymology-1 Spectroscopic techniques (structure and reactivity in solution) o Optical (circular dichroism, UV-visible, fluorescence) o Vibrational (infrared, Raman) Electrochemical methods (kinetic analysis) o Potentiometric techniques o Conductometry Enthalpimetry (microcalorimetry) o Very sensitive and free of interference Radiochemical methods o Far more sensitive than photometric ones but. . .

Tools of enzymology-2 X-ray crystallography o First crystallized enzyme, urease (J. Sumner, in 1926) crystals are proteins and their dissolution led to enzymatic activity o Within 20 years: >130 enzymes crystals documented o 3 -D structure of a protein, myoglobin, was deduced (Kendrew, 1957) Multidimentional nuclear magnetic resonance (NMR) and X-ray crystallography are now commonly used: n n to explain the mechanistic details of enzyme catalysis to design new ligands Molecular Biology o Clone and express enzymes in foreign hosts (overexpression purification and characterization of enzymes occuring naturally in minute quantity) o Manipulate the a. acid sequence (site-directed mutagenesis and deletional mutagenesis chemical groups in ligand binding)

The effect of enzyme inhibition Irreversible inhibitors: Combine with the functional groups of the amino acids in the active site, irreversibly. Examples: nerve gases and pesticides, containing organophosphorus, combine with serine residues in the enzyme acetylcholine esterase. o © 2007 Paul Billiet ODWS

The effect of enzyme inhibition Reversible inhibitors: These can be washed out of the solution of enzyme by dialysis. There are two categories. o © 2007 Paul Billiet ODWS

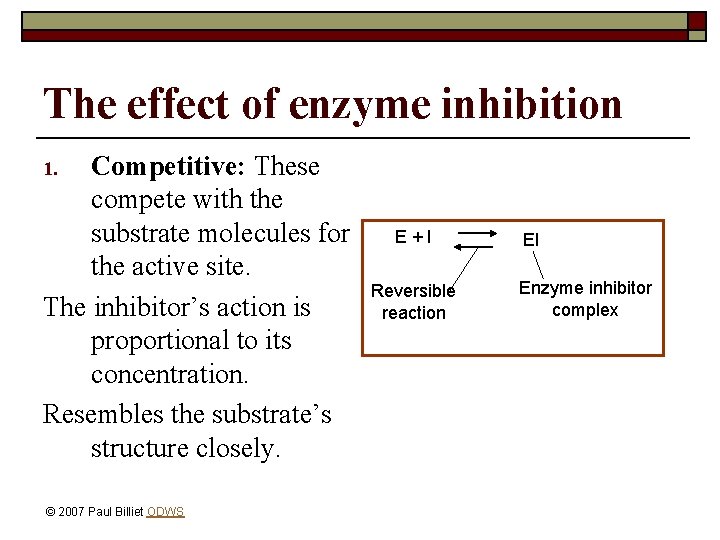
The effect of enzyme inhibition Competitive: These compete with the substrate molecules for the active site. The inhibitor’s action is proportional to its concentration. Resembles the substrate’s structure closely. 1. © 2007 Paul Billiet ODWS E+I Reversible reaction EI Enzyme inhibitor complex

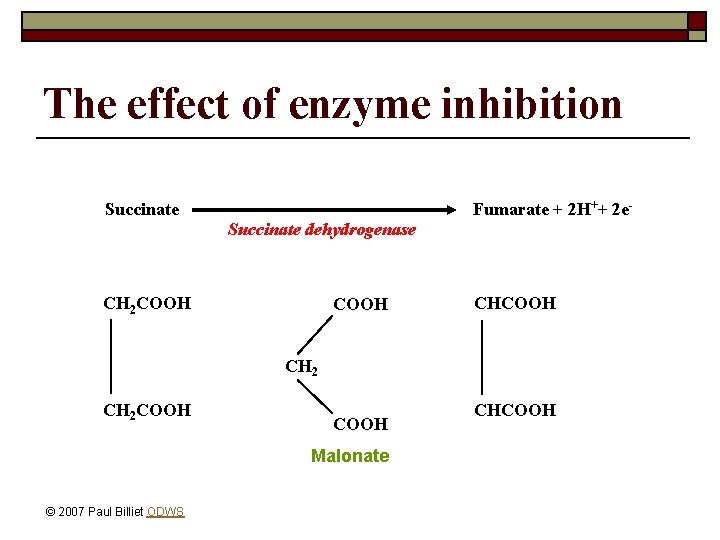
The effect of enzyme inhibition Fumarate + 2 H++ 2 e- Succinate dehydrogenase CH 2 COOH CHCOOH CH 2 COOH Malonate © 2007 Paul Billiet ODWS CHCOOH

The effect of enzyme inhibition Non-competitive: These are not influenced by the concentration of the substrate. It inhibits by binding irreversibly to the enzyme but not at the active site. Examples o Cyanide combines with the Iron in the enzymes cytochrome oxidase. o Heavy metals, Ag or Hg, combine with –SH groups. These can be removed by using a chelating agent such as EDTA. 2. © 2007 Paul Billiet ODWS

Applications of inhibitors o o o Negative feedback: end point or end product inhibition Poisons snake bite, plant alkaloids and nerve gases. Medicine antibiotics, sulphonamides, sedatives and stimulants © 2007 Paul Billiet ODWS
 Chapter 12 enzymes the protein catalyst
Chapter 12 enzymes the protein catalyst Protein that acts as biological catalyst
Protein that acts as biological catalyst Four properties of enzymes
Four properties of enzymes Protein pump vs protein channel
Protein pump vs protein channel Protein-protein docking
Protein-protein docking Proteins with catalytic power are called
Proteins with catalytic power are called Catalytic converter reaction
Catalytic converter reaction Catalytic functions
Catalytic functions Catalytic air purifier
Catalytic air purifier Catalytic converter
Catalytic converter Emery oil
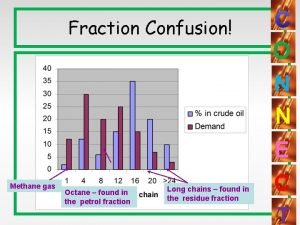
Emery oil Catalytic cracking
Catalytic cracking Cracking of octane
Cracking of octane Catalytic heater oil and gas
Catalytic heater oil and gas Site:slidetodoc.com
Site:slidetodoc.com Catalytic reforming of hexane
Catalytic reforming of hexane Naphthenes in crude oil
Naphthenes in crude oil Ethanol
Ethanol Chantal stieber
Chantal stieber Catalytic converter ingredients
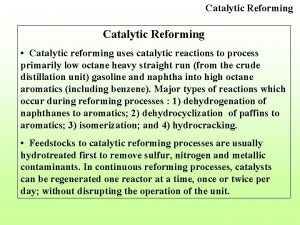
Catalytic converter ingredients Catalytic reformer bottleneck
Catalytic reformer bottleneck Figura geometrica con quattro lati e quattro angoli uguali
Figura geometrica con quattro lati e quattro angoli uguali Procedural due process vs substantive due process
Procedural due process vs substantive due process Le leggi di dracone
Le leggi di dracone Due piccole sfere identiche sono sospese a due punti
Due piccole sfere identiche sono sospese a due punti Physical properties of protein
Physical properties of protein Protein solubility
Protein solubility Protein chemical properties
Protein chemical properties Due diligence checklist for oil and gas properties
Due diligence checklist for oil and gas properties Extensive vs intensive
Extensive vs intensive Chemical and physical properties
Chemical and physical properties Enzyme activators examples
Enzyme activators examples Enzymes for mayonaise
Enzymes for mayonaise Large intestine function in digestive system
Large intestine function in digestive system