DREAM 6 Flow CAP 2 Challenge Molecular Classification


![DREAM 6/Flow. CAP 2 challenge 2011 The DREAM project [www. the-dream-project. org] Dialogue for DREAM 6/Flow. CAP 2 challenge 2011 The DREAM project [www. the-dream-project. org] Dialogue for](https://slidetodoc.com/presentation_image_h/13122f709ab3129797ed77a19b30b59b/image-3.jpg)











- Slides: 25

DREAM 6 / Flow. CAP 2 Challenge: Molecular Classification of Acute Myeloid Leukaemia Team Admire-LVQ Adaptive Distance Measures In Relevance Learning Vector Quantization Michael Biehl Kerstin Bunte Petra Schneider Johann Bernoulli Institute for Mathematics and Computer Science University of Groningen, The Netherlands Centre for Diabetes, Endicronology & Metabolism School of Clinical & Experimental Medicine University of Birmingham, UK 1

![DREAM 6Flow CAP 2 challenge 2011 The DREAM project www thedreamproject org Dialogue for DREAM 6/Flow. CAP 2 challenge 2011 The DREAM project [www. the-dream-project. org] Dialogue for](https://slidetodoc.com/presentation_image_h/13122f709ab3129797ed77a19b30b59b/image-3.jpg)
DREAM 6/Flow. CAP 2 challenge 2011 The DREAM project [www. the-dream-project. org] Dialogue for Reverse Engineering Assessments and Methods Organizers Gustavo Stolovitzky, Robert Prill, Raquel Norel, Pablo Meyer, IBM Computational Biology Center Julio Saez-Rodriguez, European Bioinformatics Institute (EMBL-EBI) Flow. CAP initiative [http: //flowcap. flowsite. org] Flow Cytometry: Critical Assessment of Population Identification Methods Organizers Ryan Brinkman, British Columbia Cancer Agency Raphael Gottardo, Fred Hutchinson Cancer Research Center Tim Mosmann, University of Rochester Richard H. Scheuermann, University of Texas Southwestern Medical Center 3

flow cytometry peripheral blood/ bone marrow aspirate preprocessing fluorophoreconjugated antibodies for specific proteins cell size, granularity, +26 protein markers (ten-) thousands of events per marker training set: 23 AML patients, 156 healthy donors Wade Rogers, test set U. of Pennsylvania : 180 unlabeled patients © www. the-dream-project. org 4

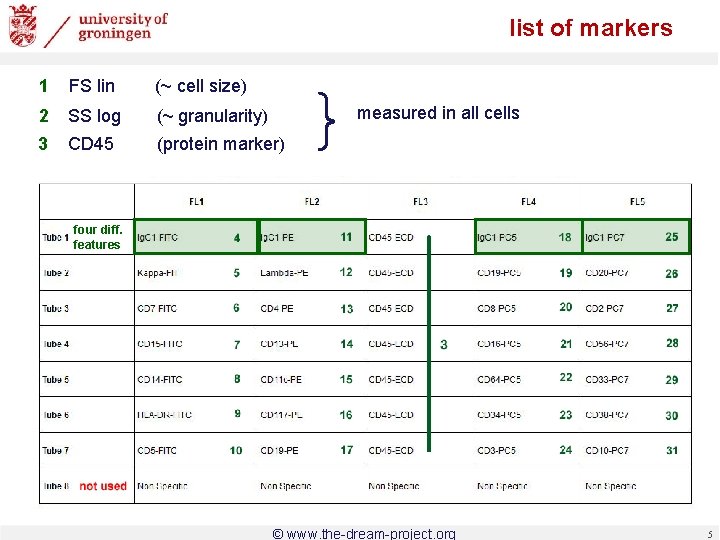
list of markers 1 FS lin (~ cell size) 2 SS log (~ granularity) 3 CD 45 (protein marker) } measured in all cells four diff. features © www. the-dream-project. org 5

list of markers possible workflow: - selection of cells, based on e. g. FS Lin, SS Log, CD-45 - inspection of all markers only for selected cells e. g. differential diagnosis (subtypes) here: classification based on entire cell population and all markers target diagnosis: AML patient / healthy donor unspecific with respect to types of AML consideration of frequencies / histograms only information about single cells disregarded 6

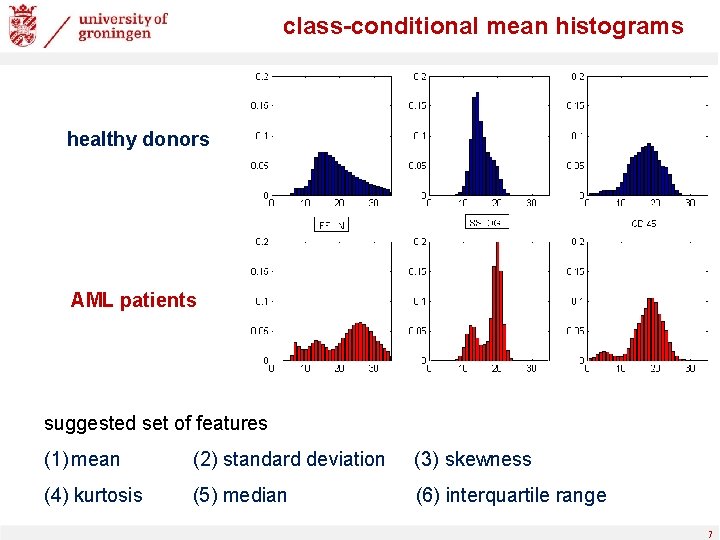
class-conditional mean histograms healthy donors AML patients suggested set of features (1) mean (2) standard deviation (3) skewness (4) kurtosis (5) median (6) interquartile range 7

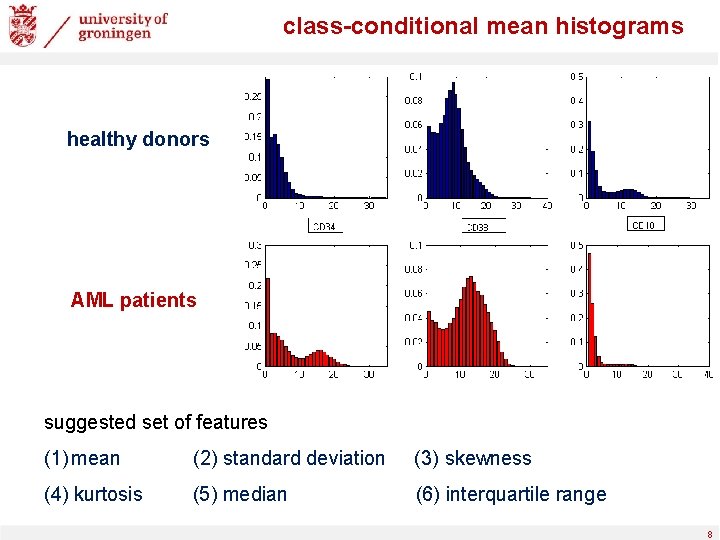
class-conditional mean histograms healthy donors AML patients suggested set of features (1) mean (2) standard deviation (3) skewness (4) kurtosis (5) median (6) interquartile range 8

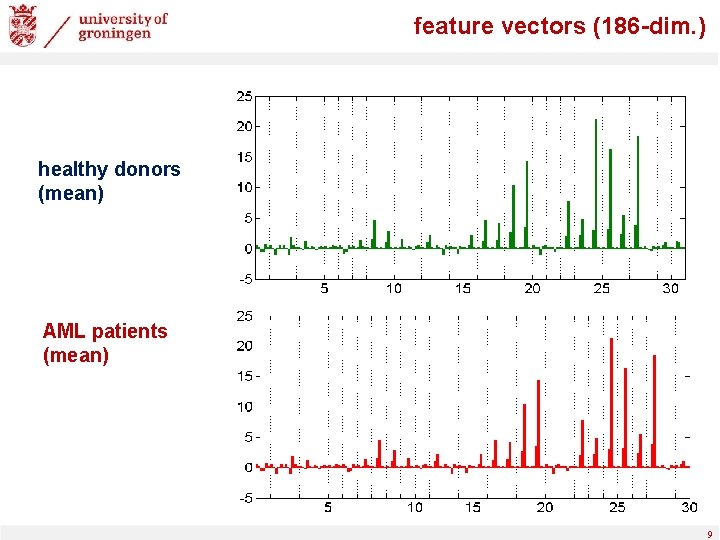
feature vectors (186 -dim. ) healthy donors (mean) AML patients (mean) 9

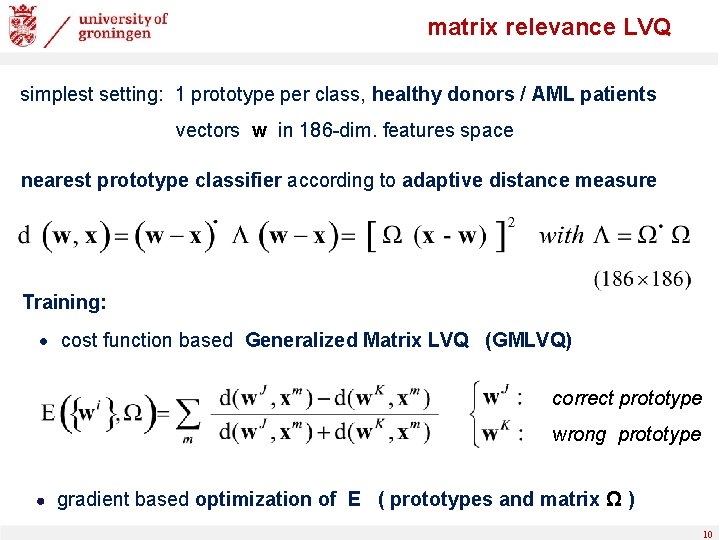
matrix relevance LVQ simplest setting: 1 prototype per class, healthy donors / AML patients vectors w in 186 -dim. features space nearest prototype classifier according to adaptive distance measure Training: ∙ cost function based Generalized Matrix LVQ (GMLVQ) correct prototype wrong prototype ∙ gradient based optimization of E ( prototypes and matrix Ω ) 10

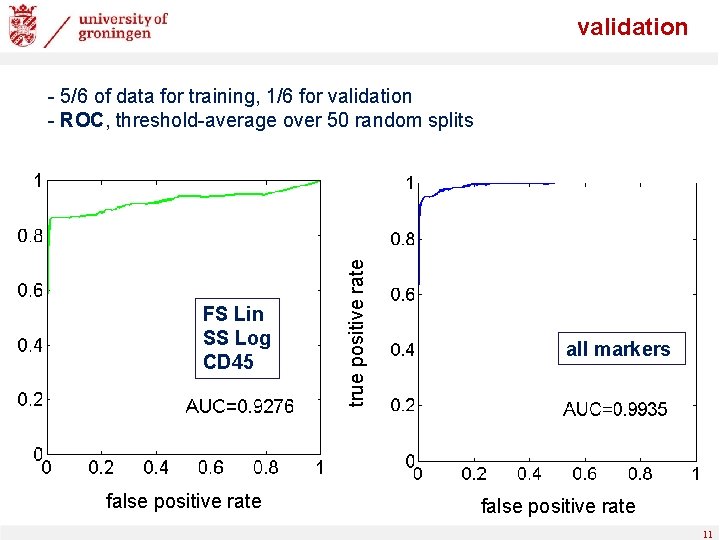
validation FS Lin SS Log CD 45 false positive rate true positive rate - 5/6 of data for training, 1/6 for validation - ROC, threshold-average over 50 random splits all markers false positive rate 11

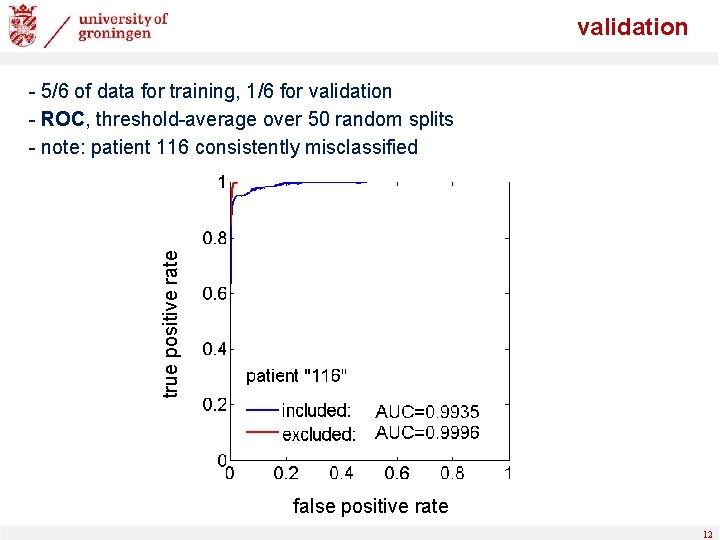
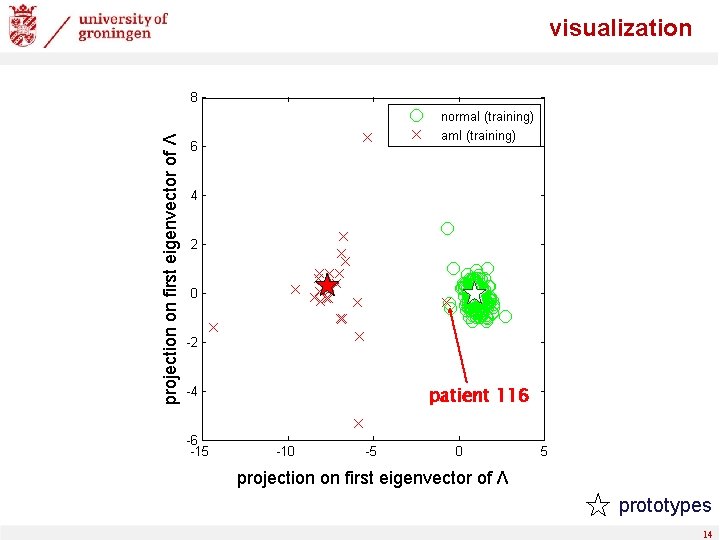
validation true positive rate - 5/6 of data for training, 1/6 for validation - ROC, threshold-average over 50 random splits - note: patient 116 consistently misclassified false positive rate 12

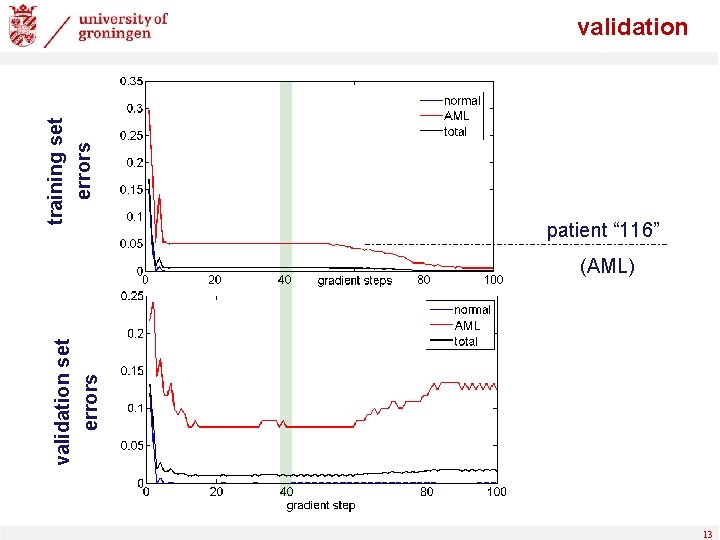
validation set errors training set errors validation patient “ 116” (AML) 13

projection on first eigenvector of Λ visualization patient 116 projection on first eigenvector of Λ prototypes 14

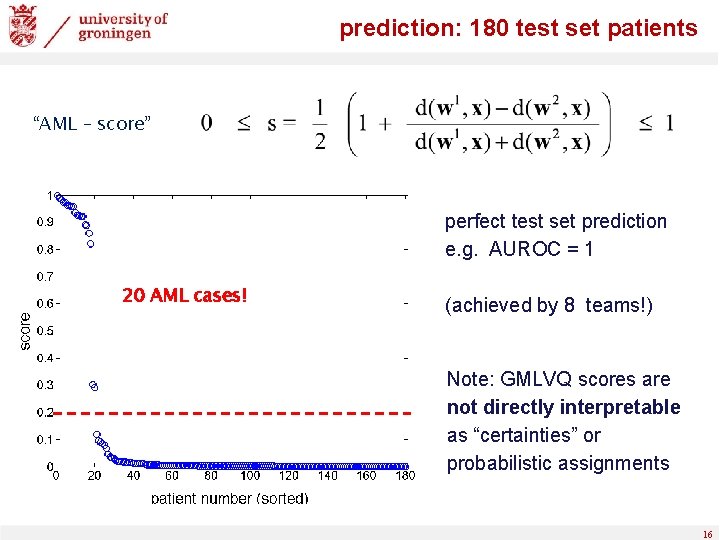
projection on first eigenvector of Λ prediction: 180 test set patients test set projection on first eigenvector of Λ prototypes 15

prediction: 180 test set patients “AML – score” perfect test set prediction e. g. AUROC = 1 20 AML cases! (achieved by 8 teams!) Note: GMLVQ scores are not directly interpretable as “certainties” or probabilistic assignments 16

prototypes difference vector “ AML - healthy ” prototype here: components corresponding to mean values 17

relevances relevance of markers: in detail: iqr median kurtosis skewness std. dev. mean ← diagonal elements of Λ 18

relevances relevance of markers: in detail: SS log iqr median kurtosis skewness std. dev. mean 19

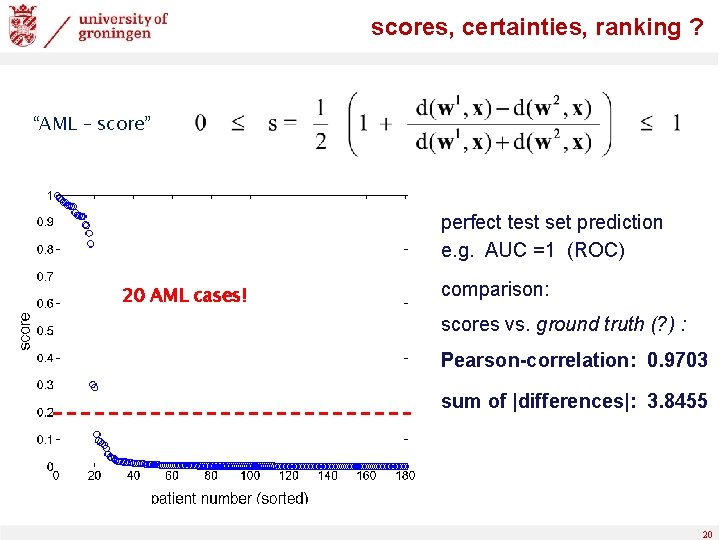
scores, certainties, ranking ? “AML – score” perfect test set prediction e. g. AUC =1 (ROC) 20 AML cases! comparison: scores vs. ground truth (? ) : Pearson-correlation: 0. 9703 sum of |differences|: 3. 8455 20

scores, certainties, ranking ? “transformed AML – score” 20 AML cases! perfect test set prediction e. g. AUC =1 (ROC) comparison: scores vs. ground truth: Pearson-correlation: 0. 9820 sum of |differences|: 4. 4347 Pearson-correlation: 0. 9703 sum of |differences|: 3. 8455 21

summary feature vectors: moment based characteristics of flow cytometry data [mean, standard deviation, skewness, kurtosis, median, iqr ] Matrix Relevance Learning Vector Quantization - perfect classification with respect to training and test set (e. g. AUC(roc)=1) - weighting of features (pairs of features) according to their relevance in the classification - visualization of the data set - identification of outliers (“ 116” ? ) 22

outlook selection of reduced feature set: relevance matrix results suggest a selection of protein markers and/or specific features direct classification of histograms non-Euclidean, histogram-specific distance measures e. g. Divergence-based LVQ [Mwebaze et al. , 2010] identification / diagnosis of AML subtypes - AML subtypes to be identified by specific marker profiles - machine learning approach requires larger data sets, e. g. GMLVQ with several prototypes representing AML - back to gating – selection of cells for differential diagnosis? 23

references (www. cs. rug. nl/~biehl) The method (GMLVQ): P. Schneider, M. Biehl, B. Hammer, Adaptive relevance matrices in learning vector quantization Neural Computation 21: 3532 -3561 (2009) A recent application in tumor classification: W. Arlt, M. Biehl, A. E. Taylor et al. J Clinical Endocrinology & Metabolism, in press (2011) Urine Steroid Metabolomics as a Biomarker Tool for Detecting Malignancy in Patients with Adrenal Tumors 24

thanks Thanks 25