Draft genome of Cicer reticulatum L the wild


- Slides: 17

Draft genome of Cicer reticulatum L. , the wild progenitor of chickpea provides insights into the domestication process SUPPORTING FIGURES

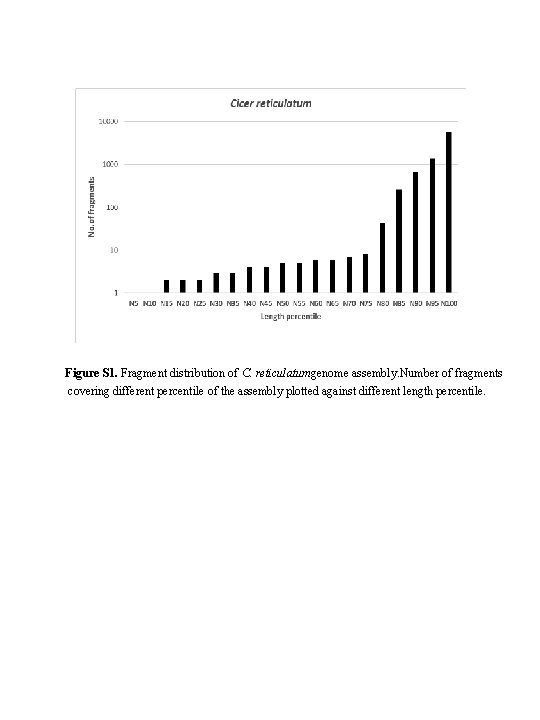
Figure S 1. Fragment distribution of C. reticulatumgenome assembly. Number of fragments covering different percentile of the assembly plotted against different length percentile.

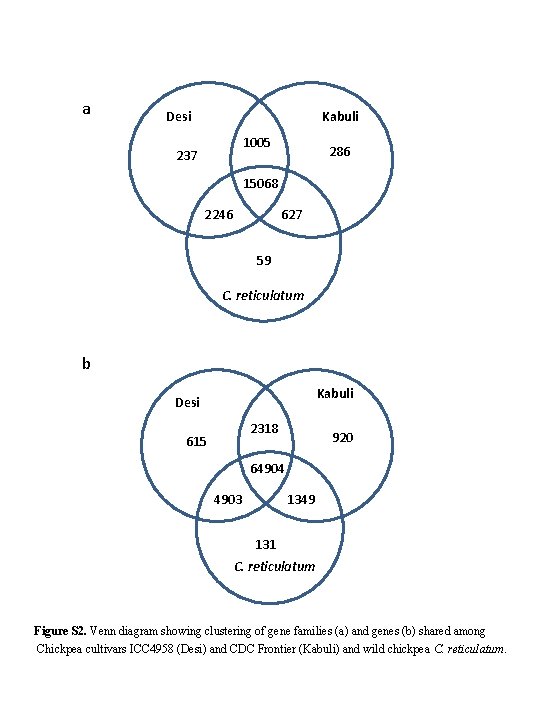
a Desi Kabuli 1005 237 286 15068 2246 627 59 C. reticulatum b Kabuli Desi 2318 615 920 64904 4903 1349 131 C. reticulatum Figure S 2. Venn diagram showing clustering of gene families (a) and genes (b) shared among Chickpea cultivars ICC 4958 (Desi) and CDC Frontier (Kabuli) and wild chickpea C. reticulatum.

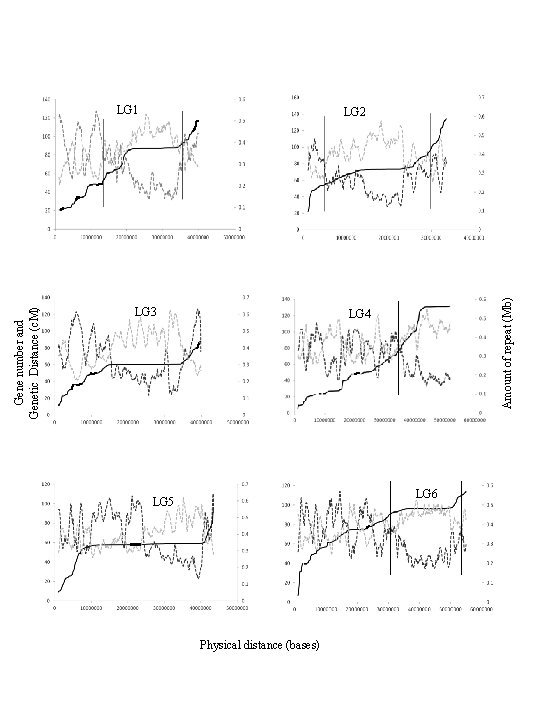
Gene number and Genetic Distance (c. M) LG 3 LG 4 LG 5 Physical distance (bases) Amount of repeat (Mb) LG 1 LG 2 LG 6

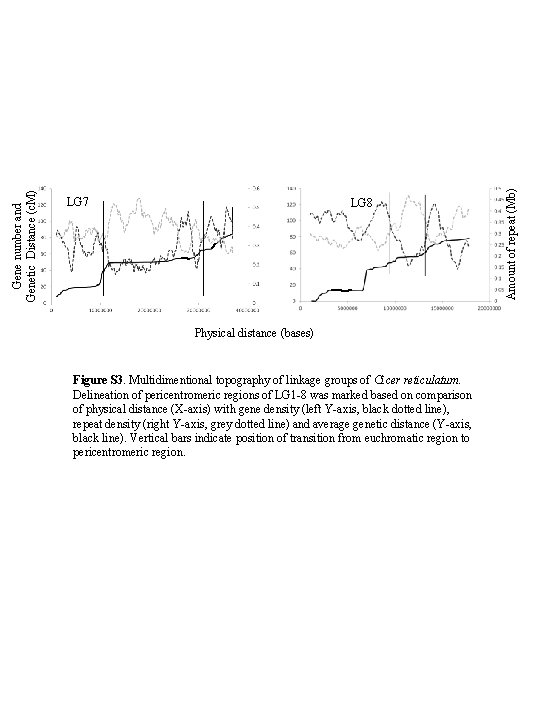
LG 8 Physical distance (bases) Figure S 3. Multidimentional topography of linkage groups of Cicer reticulatum. Delineation of pericentromeric regions of LG 1 -8 was marked based on comparison of physical distance (X-axis) with gene density (left Y-axis, black dotted line), repeat density (right Y-axis, grey dotted line) and average genetic distance (Y-axis, black line). Vertical bars indicate position of transition from euchromatic region to pericentromeric region. Amount of repeat (Mb) Gene number and Genetic Distance (c. M) LG 7

C. reticulatum C. arietinum ICC 4958 Figure S 4. Comparison of C. reticulatum and C. arietinum ICC 4958 chickpea genome assemblies. A dot-plot matrix comparing the chickpea (ICC 4958) and the C. reticulatum draft assemblies of eight pseudomolecules corresponding to each linkage groups. Pairwise comparison of all the pseudomolecules two draft genome assemblies were performed using synteny blocks and anchor filtering algorithms of tool Sy. Map v 4. 0.

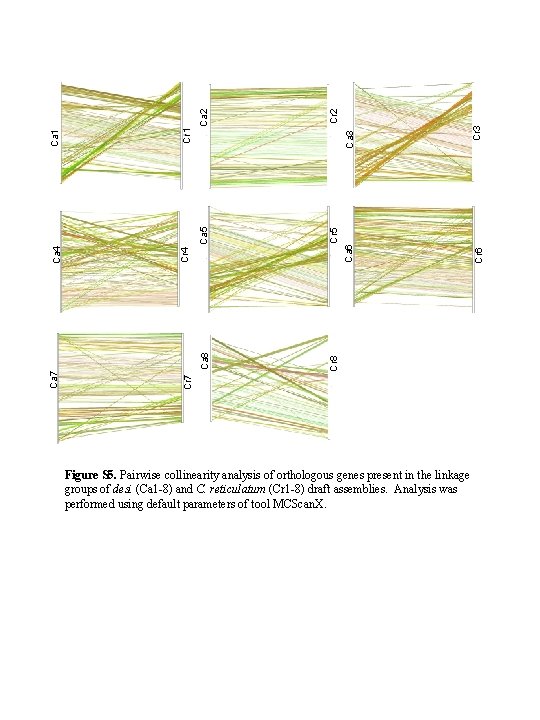
Ca 3 Ca 6 Cr 3 Cr 2 Cr 5 Cr 8 Cr 6 Ca 2 Ca 5 Ca 8 Cr 1 Cr 4 Ca 1 Ca 4 Ca 7 Cr 7 Figure S 5. Pairwise collinearity analysis of orthologous genes present in the linkage groups of desi (Ca 1 -8) and C. reticulatum (Cr 1 -8) draft assemblies. Analysis was performed using default parameters of tool MCScan. X.

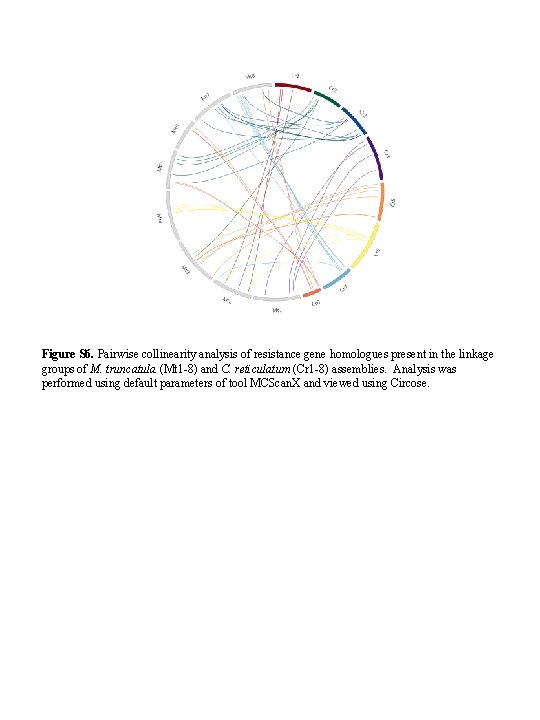
Figure S 6. Pairwise collinearity analysis of resistance gene homologues present in the linkage groups of M. truncatula (Mt 1 -8) and C. reticulatum (Cr 1 -8) assemblies. Analysis was performed using default parameters of tool MCScan. X and viewed using Circose.

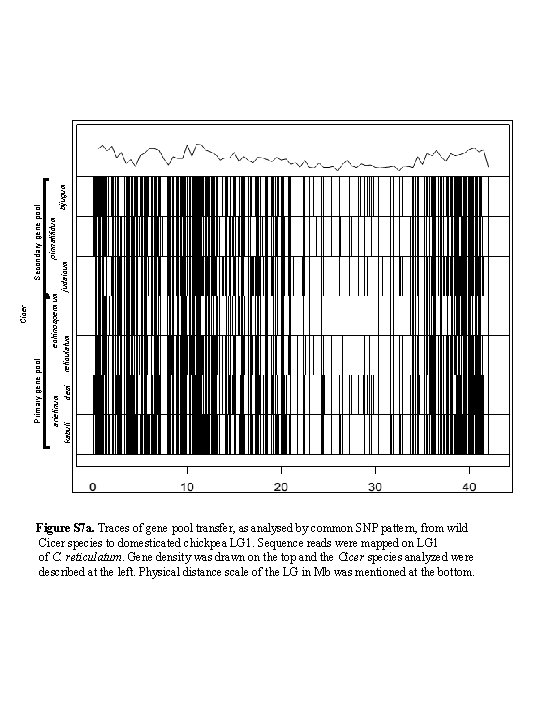
pinnatifidum echinospermum bijugum judaicum reticulatum arietinum kabuli desi Secondary gene pool Cicer Primary gene pool Figure S 7 a. Traces of gene pool transfer, as analysed by common SNP pattern, from wild Cicer species to domesticated chickpea LG 1. Sequence reads were mapped on LG 1 of C. reticulatum. Gene density was drawn on the top and the Cicer species analyzed were described at the left. Physical distance scale of the LG in Mb was mentioned at the bottom.

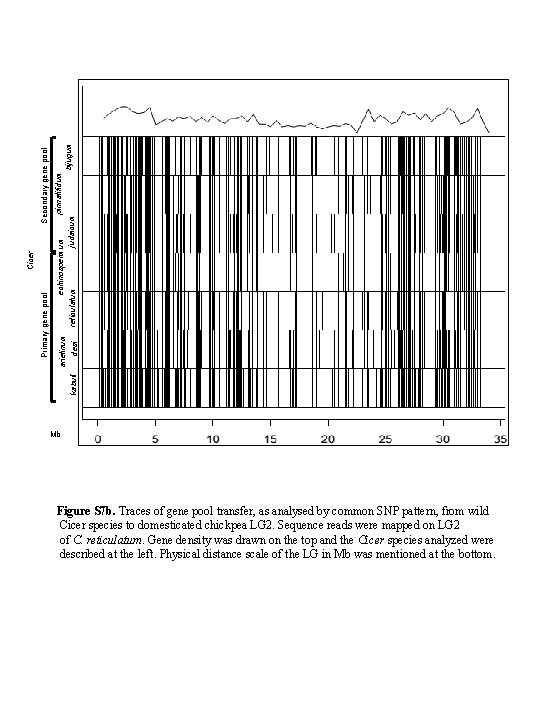
Secondary gene pool Cicer Primary gene pool pinnatifidum echinospermum arietinum bijugum judaicum reticulatum kabuli desi Mb Figure S 7 b. Traces of gene pool transfer, as analysed by common SNP pattern, from wild Cicer species to domesticated chickpea LG 2. Sequence reads were mapped on LG 2 of C. reticulatum. Gene density was drawn on the top and the Cicer species analyzed were described at the left. Physical distance scale of the LG in Mb was mentioned at the bottom.

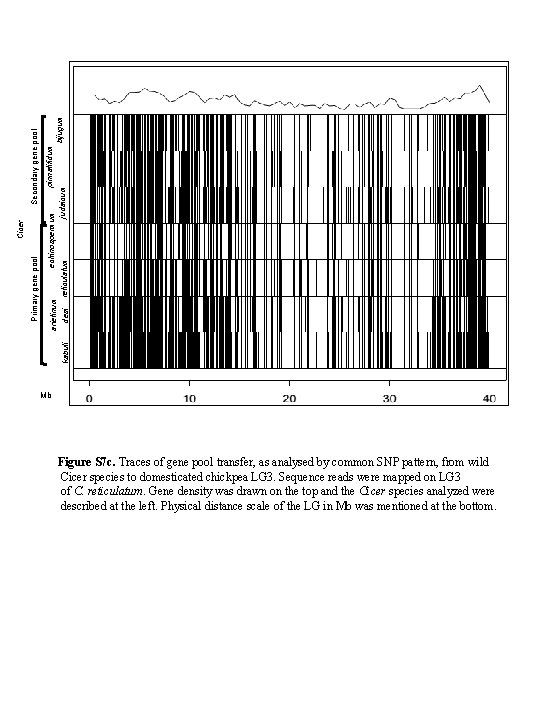
Secondary gene pool Primary gene pool Cicer pinnatifidum echinospermum arietinum bijugum judaicum desi reticulatum kabuli Mb Figure S 7 c. Traces of gene pool transfer, as analysed by common SNP pattern, from wild Cicer species to domesticated chickpea LG 3. Sequence reads were mapped on LG 3 of C. reticulatum. Gene density was drawn on the top and the Cicer species analyzed were described at the left. Physical distance scale of the LG in Mb was mentioned at the bottom.

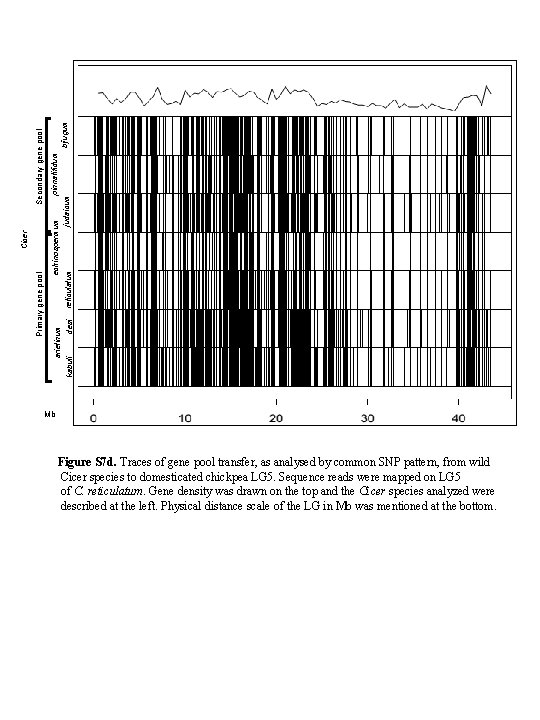
pinnatifidum echinospermum bijugum judaicum reticulatum Secondary gene pool Cicer Primary gene pool arietinum kabuli desi Mb Figure S 7 d. Traces of gene pool transfer, as analysed by common SNP pattern, from wild Cicer species to domesticated chickpea LG 5. Sequence reads were mapped on LG 5 of C. reticulatum. Gene density was drawn on the top and the Cicer species analyzed were described at the left. Physical distance scale of the LG in Mb was mentioned at the bottom.

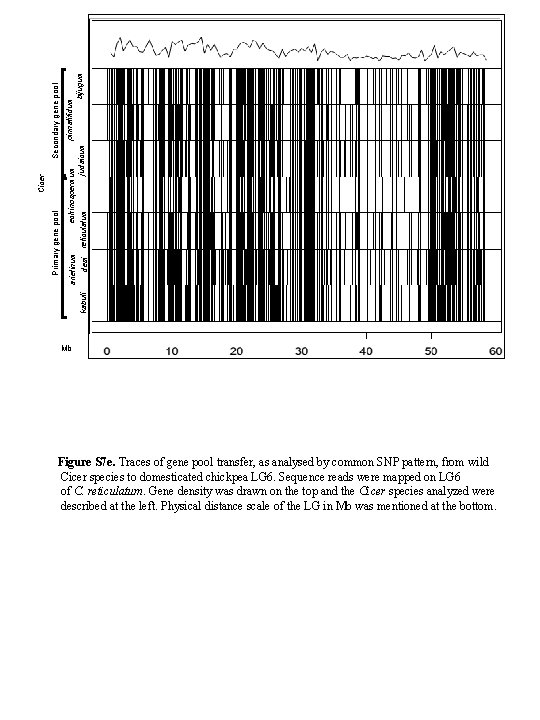
pinnatifidum echinospermum arietinum bijugum judaicum kabuli desi reticulatum Secondary gene pool Cicer Primary gene pool Mb Figure S 7 e. Traces of gene pool transfer, as analysed by common SNP pattern, from wild Cicer species to domesticated chickpea LG 6. Sequence reads were mapped on LG 6 of C. reticulatum. Gene density was drawn on the top and the Cicer species analyzed were described at the left. Physical distance scale of the LG in Mb was mentioned at the bottom.

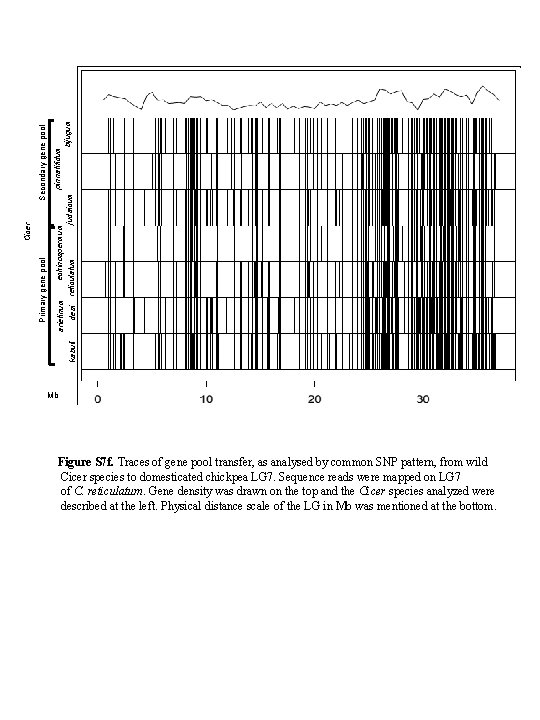
kabuli desi pinnatifidum echinospermum bijugum judaicum reticulatum Secondary gene pool Cicer Primary gene pool arietinum Mb Figure S 7 f. Traces of gene pool transfer, as analysed by common SNP pattern, from wild Cicer species to domesticated chickpea LG 7. Sequence reads were mapped on LG 7 of C. reticulatum. Gene density was drawn on the top and the Cicer species analyzed were described at the left. Physical distance scale of the LG in Mb was mentioned at the bottom.

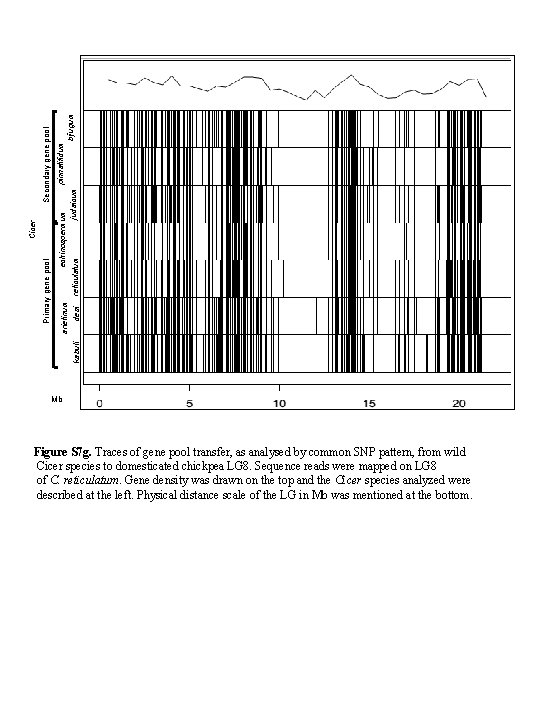
kabuli pinnatifidum echinospermum arietinum bijugum judaicum desi reticulatum Secondary gene pool Primary gene pool Cicer Mb Figure S 7 g. Traces of gene pool transfer, as analysed by common SNP pattern, from wild Cicer species to domesticated chickpea LG 8. Sequence reads were mapped on LG 8 of C. reticulatum. Gene density was drawn on the top and the Cicer species analyzed were described at the left. Physical distance scale of the LG in Mb was mentioned at the bottom.

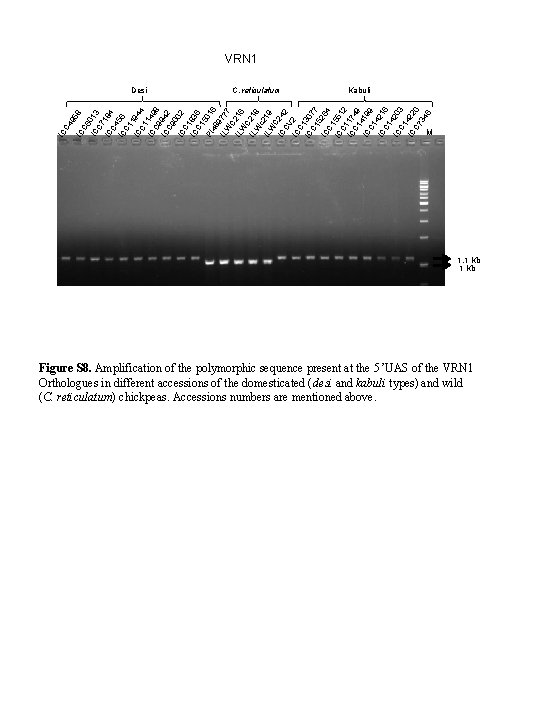
VRN 1 C 4 IC C. reticulatum Kabuli 9 IC 58 C 6 IC 013 C 7 IC 184 C 4 IC 56 C 1 1 IC 94 C 1 4 IC 149 C 9 8 IC 942 C 9 0 IC 02 C 1 IC 836 C 1 5 PI 016 48 9 IL 77 7 W C 2 IL 1 W 6 C IL 218 W c IL 219 W C IC 242 CV IC 2 C 1 IC 307 C 1 7 5 IC 264 C 1 IC 551 C 1 2 IC 17 C 1 49 IC 419 C 1 9 IC 421 C 1 6 4 IC 203 C 1 IC 422 C 7 0 34 6 Desi M 1. 1 Kb Figure S 8. Amplification of the polymorphic sequence present at the 5 ’UAS of the VRN 1 Orthologues in different accessions of the domesticated (desi and kabuli types) and wild (C. reticulatum) chickpeas. Accessions numbers are mentioned above.

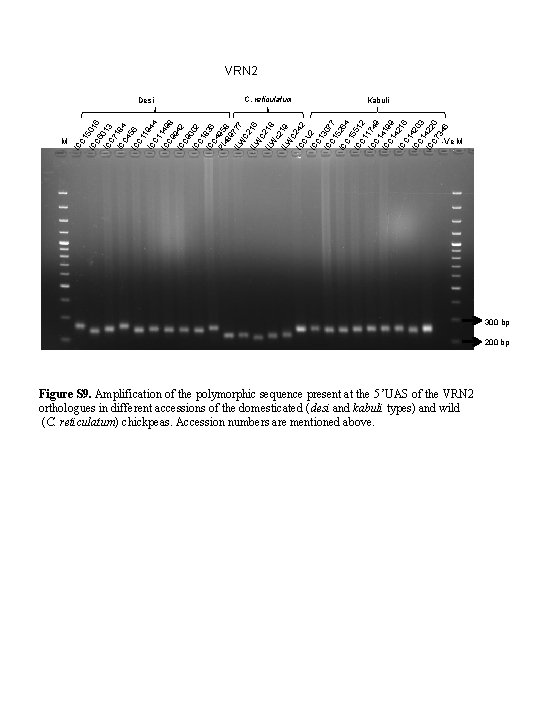
VRN 2 IC M C. reticulatum Kabuli C 1 IC 501 C 6 6 IC 013 C 7 IC 184 C 4 IC 56 C 1 1 IC 944 C 1 IC 149 C 9 8 9 IC 42 C 9 IC 002 C 1 IC 836 C 4 PI 95 48 8 9 IL 777 W C 2 1 IL W 6 C 2 1 IL W 8 c 2 1 IL W 9 C IC 242 CV IC 2 C 1 IC 307 C 1 7 5 IC 264 C 1 IC 551 C 1 2 IC 174 C 1 9 IC 419 C 1 9 4 IC 216 C 1 IC 420 C 1 3 IC 422 C 7 0 34 6 Desi -Ve M 300 bp 200 bp Figure S 9. Amplification of the polymorphic sequence present at the 5 ’UAS of the VRN 2 orthologues in different accessions of the domesticated (desi and kabuli types) and wild (C. reticulatum) chickpeas. Accession numbers are mentioned above.
 Kelas ophioglossum reticulatum
Kelas ophioglossum reticulatum Semi-global alignment
Semi-global alignment Ladislav cicer
Ladislav cicer Heel and toe arrangement in carding
Heel and toe arrangement in carding Wild born
Wild born Wild sweet pea vs wild potato
Wild sweet pea vs wild potato National human genome research institute
National human genome research institute Human genome structure
Human genome structure Genome definition
Genome definition Future of human genome project
Future of human genome project Genome mapping
Genome mapping What is genome
What is genome Genome.gov
Genome.gov Genome identification
Genome identification Genome klick
Genome klick Chapter 14 the human genome
Chapter 14 the human genome Human genome project source code
Human genome project source code Broad institute igv
Broad institute igv